International Journal of Clinical Endocrinology and Metabolism
Investigating polymorphisms in genes encoding TGF-β1, IL-10, and IL-6 and their associations with type 1 diabetes mellitus
Ahmed H Alghamdi1, Mohamed F El-Refaei1,2*, Ibrahim M Shatla1,3 and Sherif M El-Sherbini2,4
2Genetic Institute, Sadat City University, Egypt
3Demietta Faculty of Medicine, Al-Azhar University, Egypt
4Faculty of Science, Al-Baha University, Al-Baha, Saudi Arabia
Cite this as
Alghamdi AH, El-Refaei MF, Shatla IM, El-Sherbini SM (2023) Investigating polymorphisms in genes encoding TGF-β1, IL-10, and IL-6 and their associations with type 1 diabetes mellitus. Int J Clin Endocrinol Metab 9(1): 019-025. DOI: 10.17352/ijcem.000058Copyright License
© 2023 Alghamdi AH, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Many cytokines play a role in the pathogenesis of Type 1 Diabetes (T1D), and gene polymorphisms could possibly contribute to the disease’s genetic predisposition because they can affect cytokine production or function. The purpose of this study was to investigate the role of the gene polymorphisms TGF-β1 (+869T/C), (+915G/C), IL-10 {-1082 G/A), (-819 C/T), and (-592 A/C), and IL-6 (-174 G/C) in hereditary vulnerability to T1D. The Polymerase Chain Reaction with Sequence-Specific Primers (PCR-SSP) was used to analyze the polymorphisms. According to their genotypes, individuals were divided into the low-, high-, or intermediate-producer phenotypes predicted for these cytokines polymorphisms. Our findings revealed that the production of TGF-β1 was significantly higher in control than in T1D participants whereas the IL-6 genotype with low IL-6 production was significantly increased in the cases compared to the control. A significant association was evident between TGF-β1 and IL-6 low production and the incidence of T1D, thereby confirming the importance of TGF-β1 and IL-6 polymorphism as a genetic factor contributing to the incidence of T1D. By contrast, the involvement of IL-10 in the incidence of T1D was not as clear. Although some evidence supports a relationship, no statistically significant association has been verified between IL-10 and T1D. This type of measurement could be beneficial in determining the susceptibility and severity of the T1D condition while also taking into consideration the prediction of T1D incidence.
Introduction
The autoimmune condition known as Type 1 Diabetes (T1D) is mediated by T cells that specifically kill cells that produce insulin [1]. The activation and expansion of autoreactive CD4+ T cells and CD8+ cytotoxic T cells are thought to be facilitated by B cells, an important class of antigen-presenting cells that express costimulatory signaling molecules and are implicated in the development of T1D [2]. The collapse of the immune system is primarily mediated by T helper 1 (Th1) cells. Whereas islet infiltration, immune cell activation, and all other mediators contribute to the destruction of pancreatic cells and the overt hyperglycemia observed in this disease [3]. Exogenous insulin replacement is the mainstay of current T1D treatment, highlighting the need for specific immunotherapies to slow the progression of the condition and enhance clinical results. Genetic and immunopathogenic studies have directly implicated cytokines in the pathogenesis of T1D. Cytokines are the primary cause of inflammation and are essential for regulating ongoing cell degeneration [4]. Studies in mouse models, particularly in Non-Obese Diabetic (NOD) mice, a recognized animal model of T1D, have demonstrated that the modulation of cytokine function can be a therapeutic strategy, and a number of novel cytokines are now identified as potential therapeutic targets for combating immune-mediated cell damage [5]. For these uses, cytokines are categorized into three classes: those with conventionally anti-inflammatory roles (e.g., IL-10 and TGF-β1 band type-2 cytokines), those with conventionally pro-inflammatory roles (e.g., IL-1, IL-6, and TNF-α), and members of the IL-12 family roles (e.g., IL-21, IL-33, [6]. However, the roles played by cytokines in the pathophysiology of T1D are currently unclear and complex, especially regarding inflammation and the course of the disease, as a large number of dysregulated cytokines become involved in the dynamics of cytokine regulation. In the context of T1D, very few cytokines have only pro- or anti-inflammatory effects. For example, a blockade of Tumor Necrosis Factor (TNF) action results in the preservation of β-cell function in children with new-onset T1D [7], while IL-2 treatment is able to increase the proportion of regulatory T cells (Tregs) without causing any negative side effects in patients with T1D [8,9]. These findings highlight the crucial role played by cytokines in T1D.
One cytokine, TGF-β1, has been associated with the control of innate and adaptive immunity and plays a significant role in many pathological and physiological responses [10-12]. The signaling sequence of the TGF-β1 protein is encoded by the TGF-β1 gene polymorphisms + 869 T/C and/or + 915 G/C, which have an impact on cytokine production [13,14]. According to several studies, the Th2 (IL-4) and Th3 (IL-10 and TGF-β1) cytokines, as well as the Tr1 and Treg cytokines and cytokine antagonists (e.g., IL-1Ra) probably have protective roles involving inhibition of the production of Th1 and pro-inflammatory cytokines [15]. Other cell types known as Bregs have recently been associated with autoimmune diseases, transplantation issues, allergies, and infections [16]. Bregs produce the inhibitory cytokine IL-10, which downregulates the immune response; therefore, Bregs are crucial for immune tolerance. The IL-10 produced by Bregs controls cell division and growth and takes part in inflammatory and immune reactions. Currently, this cytokine is considered an immunosuppressive agent [17], and Breg dysregulation is now linked to several autoimmune conditions, including Multiple Sclerosis (MS), Systemic Lupus Erythematosus (SLE), and Rheumatoid Arthritis (RA) [18,19].
The function of the pro-inflammatory cytokine IL-6 is less clear, and concrete evidence is still lacking to support any harmful or cytotoxic effect of this cytokine on pancreatic cells [20]. T cells and macrophages secrete IL-6, a multifunctional cytokine, to activate the immune system during inflammation and infection, including the inflammatory response linked to insulin resistance. A polymorphism in the 5-flanking region of the IL-6 gene on chromosome 7 at position −174 has been documented to exert an effect on its secretion and function [21]. The aim of the present study was to demonstrate the possible role of the TGF-β1 (+869T/C), (+915G/C), IL-10 (-1082 G/A), (-819 C/T), and (-592 A/C), and IL-6 (-174 G/C) polymorphisms in the incidence of T1D in Saudi children.
Methodology
T1D and control participants
Eighty children with T1D (32 males and 48 females) were gathered from Al-Baha, Saudi Arabia. 80 non-diabetic children (35 males and 45 females) without signs of autoimmune disease were enlisted as the control group. Both groups had the same level of socioeconomic and racial diversity. Patients with T1D were identified using the diagnostic criteria of the American Diabetes Association [22]. This study complied with the Ethical Committee Guidelines for Clinical Researches, and following recruitment, consent from guardians was provided for genetic analysis. Ethical approval committee of faculty of Medicine, Al-Baha University, approval number (REC/PEA/BU-FM/2023/22).
Sampling and DNA extraction
In accordance with the manufacturer’s instructions, 5 mL of venous blood was drawn into two sterile vacutainer tubes containing tri-potassium ethylene diamine tetra-acetic acid (EDTAK3); one tube was used for biochemical analysis and the other for the extraction of genomic DNA using the Wizard®Genomic DNA Purification Kit (Qiagen, Hilden, Germany) [23]. The extracted DNA was subjected to 1% agarose gel electrophoresis to verify its presence and integrity. The purity and concentration of DNA in all samples were verified using a NanoDrop instrument (Thermo Fisher Scientific Inc).
Genotyping
According to the manufacturer’s recommendations, the subjects’ genotypes were checked for the IL-6174, IL-10 1082, 819, 592, and TGF-β1+869, +915 polymorphisms using a commercially available Cytokine Genotyping Primers Kit (One Lambda®, Canoga Park, CA, USA). Individuals were categorized into the low-, high-, or intermediate-producer phenotypes predicted for these cytokines based on their genotypes, which were previously identified [24,25].
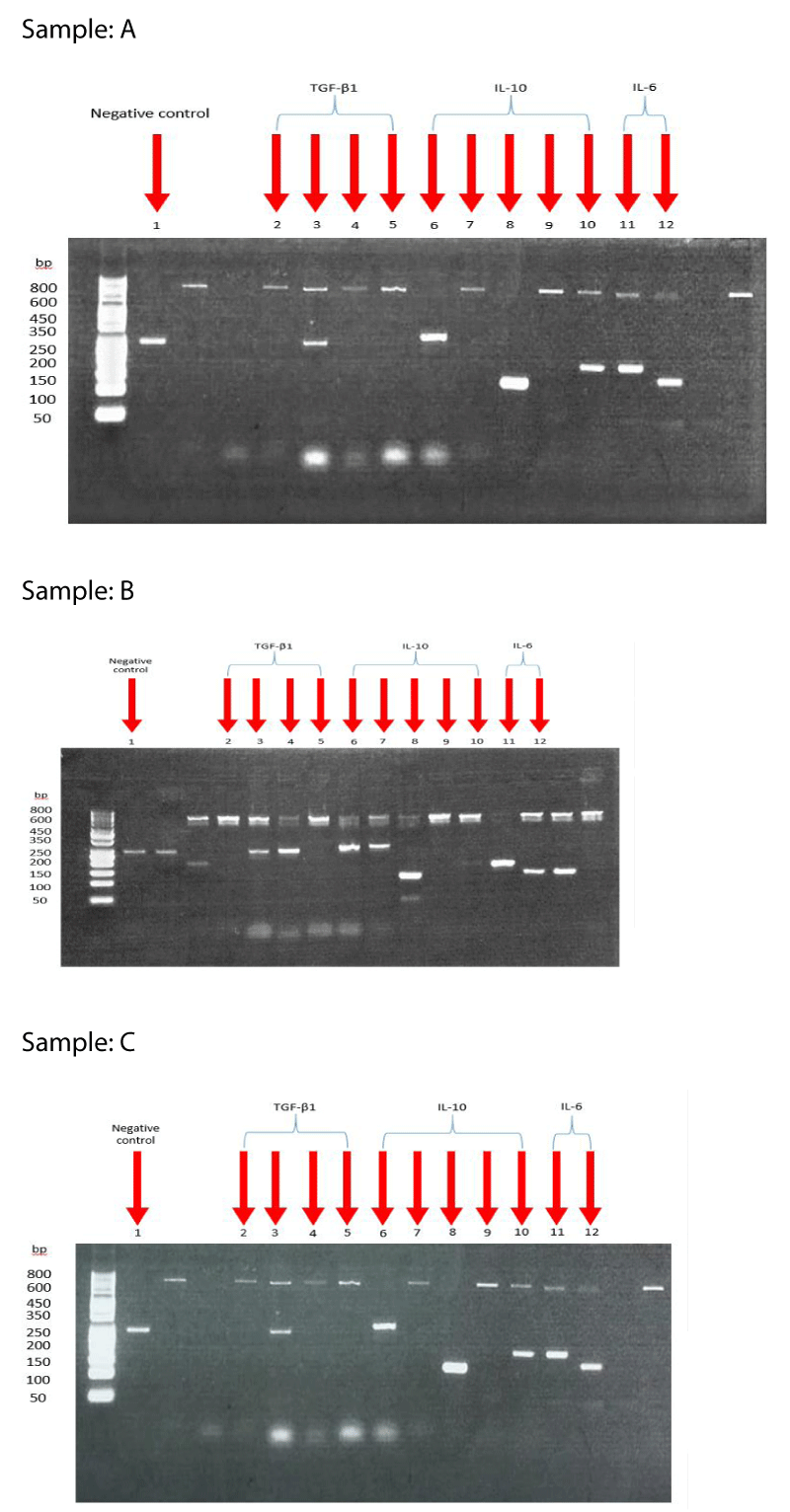
Consequently, the PCR-SSP methodology is based on the idea that completely matched oligonucleotide primers are more effectively used in amplifying a target sequence than a mismatched oligonucleotide primer by recombinant Taq polymerase than a mismatched oligonucleotide primer. Primer pairs are made to only have perfect matches with one or a small number of alleles. Perfectly matched primer pairs result in the amplification of target sequences (i.e., a positive result) under tightly controlled PCR conditions, whereas mismatched primer pairs do not (i.e., a negative result) Figure 1. Transforming growth factor-β (TGF-β), interleukin-10 (IL-10), and interleukin-6 (IL-6) genotyping were the focus of the test assay as shown in Table 1.
After being separated by agarose gel electrophoresis, the amplified DNA fragments are stained with ethidium bromide and exposed to ultraviolet light to be seen. Based on the presence or absence of a particular amplified DNA fragment, PCR-SSP results are interpreted. Thermo Fisher Scientific’s 50 bp DNA Ladder was used as a DNA ladder or marker. The gel electrophoresis image is then interpreted by using a sheet supplied by the manufacturer (WORKSHEET) to find the corresponding phenotype for the SNP of each studied gene.
Statistical analysis
The SPSS version 23 statistical program was used to code and input the data. The data were analyzed using the mean, standard deviation, and frequency for quantitative and categorical variables. The independent t-test was also used to compare data between the groups, and the chi-squared (χ2) and Fisher’s exact test were used to compare categorical variables. A probability value (p - value) less than 0.05 was considered statistically significant [26].
Results
Demographic data
Table 2 displays the demographic and biochemical details of the recruited subjects. There were 80 children participants in each study group (35 males and 45 females in the control group and 32 males and 48 females in the group of T1D patients). The age distribution of the participants with T1D and the controls was comparable. (8.6 ± 1.5 in the control group and 8.9 ± 1.9 in the patient group).
Cytokine genotype and production
The investigation of genotype and allele frequencies for the TGF-β1 (+869T/C), (+915G/C), IL-10 (-1082 G/A), (-819 C/T), and (-592 A/C), and IL-6 (-174 G/C) gene polymorphisms are listed in Table 3,4. The distribution of CC/GC and TT/GG genotypes for TGF- with intermediate and high TGF-β1 production was significantly higher in the patients with T1D than in the control group (p < 0.001). The TGF-β1 production was significantly higher in the control group than in the patient group (p < 0.001), as shown in Table 3, but no significant difference was detected in the C/T allele frequency between the patients and controls in TGF-β1 codon 10. However, a significant increase in the G allele was noted in codon 25 among control compared to patients (p < 0.001), as shown in Table 4.
Despite the presence of significant changes between the control and patients in the IL-10 genotypes but no significant changes in production. The IL-10 allele frequency showed no significant difference in the A/G frequency at position -1082, whereas a significant increase was observed at position 819 for the C allele (p < 0.0001), and at position 592 for the C allele among patients and controls (p ≤ 0.0001), as shown in Table 4. No significant difference was noted in the IL-6 genotype between the patients and the controls, also no significant difference in the G/C frequency at position -174. However, a significant increase in IL-6 with low cytokine production phenotype compared to the control was found (p < 0.0001) as shown in Tables 3,4.
Discussion
The TGF-β1 gene is polymorphic at different sites. A Single Nucleotide Polymorphism (SNP) at position +869 in the TGF-β1 promoter region causes a T-C substitution. The T allele is associated with higher concentrations of TGF β1 in plasma than is observed for the C allele, and this difference is more marked in the homozygous than in the heterozygous condition for the T allele, suggesting a gene–dose effect. Therefore, the haplotype formed by the genotypes +869 TT and +915 GG should result in the highest level of TGF-β1 synthesis, whereas the other combinations should produce a range of low- or intermediate-activity haplotypes.
The SNPs of cytokine genes are associated with both high- and low-producer phenotypes [27]; however, a few studies have found no correlation between the genotypes and the amounts of secreted cytokines [28]. Reuss, et al. [29] reported that only 50% of the observed variability in cytokine secretion could be explained by genetic factors, while environmental factors may also exert an effect.
In codon 25, the G-C substitution at position +915 causes proline to replace the expected leucine, homozygous G/G at position +915 is arg/arg. In codon 10 T-C substitution at position +869 causes proline to replace the expected arginine, homozygous T/T at position +869 is leu/leu. Higher rates of TGF-β1 production appear to be independently correlated with leucine at codon 10 and arginine at codon 25 [30,31].
Additionally, the risk of developing T1D was noticeably higher for homozygous for the codon 10 T allele than for the codon 10 C allele. TGF-β1 may prevent or delay the autoimmune-mediated destruction of pancreatic islets of Langerhans, as it is an immunosuppressive and regulatory cytokine produced by many cells, including Th3 and Treg subsets that may decrease insulin production [32,33]. TGF-β1 C/T allele at codon+869 (codon 10) did not significantly differ between T1D patients and controls, according to our findings., Also, our results revealed that one or two copies of the C allele at codon +915 (codon 25) may increase a person’s risk of developing T1D by lowering the level of the anti-inflammatory TGF-β1 as we find a significant increase in the G allele in codon 25 among control compared to patients. This finding was confirmed when investigating the cytokine gene polymorphism-associated phenotype. Moreover, a significant increase was detected in patients with low production of TGF-β1 compared to controls. Only two studies to date [34,35] have suggested that the codon 10 SNP may play a role in T1D susceptibility. Jahromi, et al. [34] discovered a significant association between the disease and the TC genotype, but not the TT genotype, in contrast, Javor, et al. [35] discovered a significant association between T1D development and the TGF-β1 codon 10 TT homozygous, but not the C allele carriers. Although both studies suggest that the TGF-β1 SNP plays a part in the propensity to develop T1D, more research is required to verify or refute this possibility.
We found no appreciable differences in genotype and phenotype between the patients and the controls for the IL-10 gene polymorphism at positions - 1082 and - 819. These findings concur with previously published findings by Reynier, et al. [36]. Also, Ide, et al. [37] reached a similar conclusion. A Japanese case study found no correlation between IL-10 gene promoter region polymorphisms and genetic susceptibility to T1D, in agreement with our findings; however, the same group in Japan also reported that only patients older than 18 years showed a significantly higher frequency of the AA genotype [38]. The IL-10 genotypes were thought to play a small role in the risk of autoimmune diabetes in Spanish T1D patients [39]. A Polish study contradicted our findings by showing an association between the IL-10-1082 polymorphism and T1D, particularly in the AA genotypes [40]. The idea that these genotypes are population-specific and may co-segregate with the disease genes in various ways among various ethnic groups may help to explain these discrepancies in findings.
Our findings contrast with those from one of the earliest case-control studies on the IL-6-174 G/C SNP, which identified GG homozygous as those at increased risk of T1D [41]. In the current study, carriers of the IL-6-174 C/C genotype were at increased risk of developing T1D. However, our findings are consistent with findings from a Polish population [42] and a sizable UK case-control study [43], which both demonstrated a marginally positive association between T1D and the -174 C allele. The pleiotropic cytokine IL-6 plays critical regulatory and pro-inflammatory roles in the pathogenesis of a number of autoimmune diseases, including rheumatoid arthritis and inflammatory bowel disease as acute inflammation is accompanied by changes in the concentrations of Acute Phase Proteins (APPs), which are controlled by IL-6 [44-46].
The role in the pathogenesis of T1D has not been established, and no concrete proof has been presented that it has harmful or even cytotoxic effects on pancreatic cells [45]. Some studies have also shown that IL-6 has a protective effect against cytokine-induced cell death and functional impairment in NOD mice, which have a genetic susceptibility to autoimmune diabetes [47,48]. Conflicting evidence has been presented regarding the association between IL-6 SNPs and T1D based on genetic studies [43,45]. The precise mechanism by which this polymorphism contributes to the genetic determination of T1D is still unclear, largely due to our incomplete understanding of the role of IL-6 in the pathogenesis of T1D and the functional impact of the -174 G/C SNP.
We found a significant increase in polymorphisms in the diabetic group’s TGF-β1 and IL-6 genes, which are linked to a noticeable alteration in the cytokine production genes. This change in cytokine production from high to low is an indication of the intricate interplay between genetic factors and the immune response. The results of our study indicate that variations in the TGF-β1 and IL-6 genes may be extremely important in determining a person’s susceptibility to Type 1 Diabetes Mellitus. Despite the fact that our findings did not find a significant correlation with IL-10 polymorphisms. The limitations of our study included the relatively small sample size, lack of diversity, and lack of direct measurements of serum cytokine levels along with gene polymorphism data that could have provided a more in-depth understanding of the mechanistic relationships between genetics, cytokine production, and disease.
Longitudinal studies that follow people over time can be used to track the development of genetic markers and how they relate to the onset of Type 1 Diabetes Mellitus. Investigate interactions between genes and the environment to understand how particular environmental elements may alter the influence of genetic variations on disease susceptibility. carrying out a thorough investigation into all genes that encode cytokines associated with Type 1 Diabetes Mellitus and their interactions with one another.
Conclusion
Our findings supported an association between susceptibility to T1D and the TGF-b1 CC/GC, TGF-β1 TT/GG. changed to TGF-β1 C/T allele at codon+869 (codon 10) did not significantly differ between T1D patients and controls while one or two copies of the C allele at codon +915 (codon 25) may increase a person’s risk of developing T1D by lowering the level of the anti-inflammatory TGF-β1. A single polymorphism, -174 C/C with low IL-6 production may be a risk factor for T1D in Saudi children. Our findings emphasize the importance of the cytokine SNPs in regulating autoimmune diseases, especially T1D, in the studied population. These results might also spur other researchers and investigators to launch further studies on larger cohorts to confirm the impact of these SNPs on the release of these cytokines and the subsequent effects on the prevalence of T1D.
Author contributions
Methodology, Ahmed H. Alghamdi, Sherif M. El-Sherbini, Ibrahim M. Shatla, and Mohamed F. El-Refaei; Investigation, Ahmed H. Alghamdi, Sherif M. El-Sherbini. Resources, Ahmed H. Alghamdi, Sherif M. El-Sherbini. Data curation, Ahmed H. Alghamdi, Sherif M. El-Sherbini, Ibrahim M. Shatla, and Mohamed F. El-Refaei.; Writing—review & editing Ahmed H. Alghamdi, Sherif M. El-Sherbini, Ibrahim M. Shatla, and Mohamed F. El-Refaei.; Visualization, Sherif M. El-Sherbini, Ibrahim M. Shatla, and Mohamed F. El-Refaei. All authors have read and agreed to the published version of the manuscript.
Funding
This study was sponsored by the Deanship of Scientific Research at Al-Baha University, Kingdom of Saudi Arabia, for their financial and logistical support and for providing necessary guidance concerning project implementation. Project No: 4/1440.
Institutional review board statement
This study complied with the Scientific Research and Ethics Committee Guidelines. Ethical approval committee of faculty of Medicine, Al-Baha University, approval number (REC/PEA/BU-FM/2023/22).
Data availability statement
The data that support the findings of this study are available on request from the corresponding author, El-Refaei, M.F. The data are not publicly available for the time being and will be available in demand.
- Gillespie KM. Type 1 diabetes: pathogenesis and prevention. CMAJ. 2006 Jul 18; 175(2):165-70. doi: 10.1503/cmaj.060244. PMID: 16847277; PMCID: PMC1489998.
- Magnuson AM, Thurber GM, Kohler RH, Weissleder R, Mathis D, Benoist C. Population dynamics of islet-infiltrating cells in autoimmune diabetes. Proc Natl Acad Sci U S A. 2015 Feb 3; 112(5):1511-6. doi: 10.1073/pnas.1423769112. Epub 2015 Jan 20. PMID: 25605891; PMCID: PMC4321317.
- DiMeglio LA, Evans-Molina C, Oram RA. Type 1 diabetes. Lancet. 2018 Jun 16; 391(10138):2449-2462. doi: 10.1016/S0140-6736(18)31320-5. PMID: 29916386; PMCID: PMC6661119.
- Nepom GT, Ehlers M, Mandrup-Poulsen T. Anti-cytokine therapies in T1D: Concepts and strategies. Clin Immunol. 2013 Dec; 149(3):279-85. doi: 10.1016/j.clim.2013.02.003. Epub 2013 Feb 14. PMID: 23510726; PMCID: PMC3700624.
- Grinberg-Bleyer Y, Baeyens A, You S, Elhage R, Fourcade G, Gregoire S, Cagnard N, Carpentier W, Tang Q, Bluestone J, Chatenoud L, Klatzmann D, Salomon BL, Piaggio E. IL-2 reverses established type 1 diabetes in NOD mice by a local effect on pancreatic regulatory T cells. J Exp Med. 2010 Aug 30; 207(9):1871-8. doi: 10.1084/jem.20100209. Epub 2010 Aug 2. PMID: 20679400; PMCID: PMC2931175.
- Mastrandrea L, Yu J, Behrens T, Buchlis J, Albini C, Fourtner S, Quattrin T. Etanercept treatment in children with new-onset type 1 diabetes: pilot randomized, placebo-controlled, double-blind study. Diabetes Care. 2009 Jul;32(7):1244-9. doi: 10.2337/dc09-0054. Epub 2009 Apr 14. PMID: 19366957; PMCID: PMC2699714.
- Hartemann A, Bensimon G, Payan CA, Jacqueminet S, Bourron O, Nicolas N, Fonfrede M, Rosenzwajg M, Bernard C, Klatzmann D. Low-dose interleukin 2 in patients with type 1 diabetes: a phase 1/2 randomised, double-blind, placebo-controlled trial. Lancet Diabetes Endocrinol. 2013 Dec; 1(4):295-305. doi: 10.1016/S2213-8587(13)70113-X. Epub 2013 Oct 8. PMID: 24622415.
- Rosenzwajg M, Churlaud G, Mallone R, Six A, Dérian N, Chaara W, Lorenzon R, Long SA, Buckner JH, Afonso G, Pham HP, Hartemann A, Yu A, Pugliese A, Malek TR, Klatzmann D. Low-dose interleukin-2 fosters a dose-dependent regulatory T cell tuned milieu in T1D patients. J Autoimmun. 2015 Apr; 58: 48-58. doi: 10.1016/j.jaut.2015.01.001. Epub 2015 Jan 26. PMID: 25634360; PMCID: PMC8153751.
- Dong S, Hiam-Galvez KJ, Mowery CT, Herold KC, Gitelman SE, Esensten JH, Liu W, Lares AP, Leinbach AS, Lee M, Nguyen V, Tamaki SJ, Tamaki W, Tamaki CM, Mehdizadeh M, Putnam AL, Spitzer MH, Ye CJ, Tang Q, Bluestone JA. The effect of low-dose IL-2 and Treg adoptive cell therapy in patients with type 1 diabetes. JCI Insight. 2021 Sep 22;6(18):e147474. doi: 10.1172/jci.insight.147474. PMID: 34324441; PMCID: PMC8492314.
- Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006; 24:99-146. doi: 10.1146/annurev.immunol.24.021605.090737. PMID: 16551245.
- You S, Thieblemont N, Alyanakian MA, Bach JF, Chatenoud L. Transforming growth factor-beta and T-cell-mediated immunoregulation in the control of autoimmune diabetes. Immunol Rev. 2006 Aug; 212:185-202. doi: 10.1111/j.0105-2896.2006.00410.x. PMID: 16903915.
- Sanjabi S, Oh SA, Li MO. Regulation of the Immune Response by TGF-β: From Conception to Autoimmunity and Infection. Cold Spring Harb Perspect Biol. 2017 Jun 1; 9(6):a022236. doi: 10.1101/cshperspect.a022236. PMID: 28108486; PMCID: PMC5453394.
- Dhaouadi T, Sfar I, Bardi R, Jendoubi-Ayed S, Abdallah TB, Ayed K, Gorgi Y. Cytokine gene polymorphisms in kidney transplantation. Transplant Proc. 2013 Jul-Aug; 45(6):2152-7. doi: 10.1016/j.transproceed.2012.12.006. Epub 2013 Jun 6. PMID: 23747182.
- Bijlsma FJ, van der Horst AA, Tilanus MG, Rozemuller E, de Jonge N, Gmelig-Meyling FH, de Weger RA. No association between transforming growth factor beta gene polymorphism and acute allograft rejection after cardiac transplantation. Transpl Immunol. 2002 Jun; 10(1):43-7. doi: 10.1016/s0966-3274(02)00021-7. PMID: 12182464.
- Wang W, Sung N, Gilman-Sachs A, Kwak-Kim J. T Helper (Th) Cell Profiles in Pregnancy and Recurrent Pregnancy Losses: Th1/Th2/Th9/Th17/Th22/Tfh Cells. Front Immunol. 2020 Aug 18; 11:2025. doi: 10.3389/fimmu.2020.02025. PMID: 32973809; PMCID: PMC7461801.
- Dasgupta S, Dasgupta S, Bandyopadhyay M. Regulatory B cells in infection, inflammation, and autoimmunity. Cell Immunol. 2020 Jun;352: 104076. doi: 10.1016/j.cellimm.2020.104076. Epub 2020 Feb 27. PMID: 32143836.
- Rosser EC, Mauri C. Regulatory B cells: origin, phenotype, and function. Immunity. 2015 Apr 21; 42(4):607-12. doi: 10.1016/j.immuni.2015.04.005. PMID: 25902480.
- Xiao S, Brooks CR, Zhu C, Wu C, Sweere JM, Petecka S, Yeste A, Quintana FJ, Ichimura T, Sobel RA, Bonventre JV, Kuchroo VK. Defect in regulatory B-cell function and development of systemic autoimmunity in T-cell Ig mucin 1 (Tim-1) mucin domain-mutant mice. Proc Natl Acad Sci U S A. 2012 Jul 24; 109(30):12105-10. doi: 10.1073/pnas.1120914109. Epub 2012 Jul 5. PMID: 22773818; PMCID: PMC3409739.
- Daien CI, Gailhac S, Mura T, Audo R, Combe B, Hahne M, Morel J. Regulatory B10 cells are decreased in patients with rheumatoid arthritis and are inversely correlated with disease activity. Arthritis Rheumatol. 2014 Aug; 66(8):2037-46. doi: 10.1002/art.38666. PMID: 24729478.
- Yin YW, Sun QQ, Zhang BB, Hu AM, Wang Q, Liu HL, Hou ZZ, Zeng YH, Xu RJ, Shi LB. The lack of association between interleukin-6 gene -174 G/C polymorphism and the risk of type 1 diabetes mellitus: a meta-analysis of 18,152 subjects. Gene. 2013 Feb 25; 515(2):461-5. doi: 10.1016/j.gene.2012.11.062. Epub 2012 Dec 13. PMID: 23246692.
- Ali YBM, El-Gahel HE, Abdel-Hakem NE, Gadalla ME, El-Hefnawy MH, El-Shahat M. Association between IL-18 and IL-6 gene polymorphisms and the risk of T1D in Egyptian children. J Diabetes Metab Disord. 2021 Mar 8; 20(1):439-446. doi: 10.1007/s40200-021-00763-w. PMID: 34222070; PMCID: PMC8212230.
- American Diabetes Association. 2. Classification and Diagnosis of Diabetes. Diabetes Care. 2016 Jan;39 Suppl 1:S13-22. doi: 10.2337/dc16-S005. Erratum in: Diabetes Care. 2016 Sep;39(9):1653. PMID: 26696675.
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988 Feb 11;16(3):1215. doi: 10.1093/nar/16.3.1215. PMID: 3344216; PMCID: PMC334765.
- Alper CM, Winther B, Hendley JO, Doyle WJ. Cytokine polymorphisms predict the frequency of otitis media as a complication of rhinovirus and RSV infections in children. Eur Arch Otorhinolaryngol. 2009 Feb;266(2):199-205. doi: 10.1007/s00405-008-0729-2. Epub 2008 Jun 17. PMID: 18560870; PMCID: PMC7087847.
- Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998 Oct 1;102(7):1369-76. doi: 10.1172/JCI2629. PMID: 9769329; PMCID: PMC508984.
- Chan YH. Biostatistics 102: quantitative data--parametric & non-parametric tests. Singapore Med J. 2003 Aug;44(8):391-6. PMID: 14700417.
- Manal Y, Tayel AN, Ibtessam M, Abdelhamid, Helmy MAS, Zaki NE, Nehad S. Elsharkawy & Amira I. Fayad TNF-α -308 G>A and IL10 -1082A>G polymorphisms as potential risk factors for lymphoproliferative disorders in autoimmune rheumatic diseases. Egyptian Journal of Medical Human Genetics 2020; 21(1). [DOI: 10.1186/s43042-019-0043-0]
- Cartwright NH, Keen LJ, Demaine AG, Hurlock NJ, McGonigle RJ, Rowe PA, Shaw JF, Szydlo RM, Kaminski ER. A study of cytokine gene polymorphisms and protein secretion in renal transplantation. Transpl Immunol. 2001 Feb;8(4):237-44. doi: 10.1016/s0966-3274(01)00026-0. PMID: 11316066.
- Reuss E, Fimmers R, Kruger A, Becker C, Rittner C, Höhler T. Differential regulation of interleukin-10 production by genetic and environmental factors--a twin study. Genes Immun. 2002 Nov;3(7):407-13. doi: 10.1038/sj.gene.6363920. PMID: 12424622.
- Li HY, Zhou T, Lin S, Lin W. Relationship between TGF-β1 + 869 T/C and + 915 G/C gene polymorphism and risk of acute rejection in renal transplantation recipients. BMC Med Genet. 2019 Jun 25;20(1):113. doi: 10.1186/s12881-019-0847-2. PMID: 31238890; PMCID: PMC6593503.
- He B, Xu C, Yang B, Landtblom AM, Fredrikson S, Hillert J. Linkage and association analysis of genes encoding cytokines and myelin proteins in multiple sclerosis. J Neuroimmunol. 1998 Jun 1;86(1):13-9. doi: 10.1016/s0165-5728(98)00003-4. PMID: 9655468.
- Ichinose K, Kawasaki E, Eguchi K. Recent advancement of understanding pathogenesis of type 1 diabetes and potential relevance to diabetic nephropathy. Am J Nephrol. 2007;27(6):554-64. doi: 10.1159/000107758. Epub 2007 Sep 6. PMID: 17823503.
- Richer MJ, Straka N, Fang D, Shanina I, Horwitz MS. Regulatory T-cells protect from type 1 diabetes after induction by coxsackievirus infection in the context of transforming growth factor-beta. Diabetes. 2008 May;57(5):1302-11. doi: 10.2337/db07-1460. Epub 2008 Feb 11. PMID: 18268045.
- Jahromi MM, Millward BA, Demaine AG. Significant correlation between association of polymorphism in codon 10 of transforming growth factor-beta1 T (29) C with type 1 diabetes and patients with nephropathy disorder. J Interferon Cytokine Res. 2010 Feb;30(2):59-66. doi: 10.1089/jir.2009.0026. PMID: 20039825.
- Javor J, Ferencik S, Bucova M, Stuchlikova M, Martinka E, Barak L, Strbova L, Grosse-Wilde H, Buc M. Polymorphisms in the genes encoding TGF-beta1, TNF-alpha, and IL-6 show association with type 1 diabetes mellitus in the Slovak population. Arch Immunol Ther Exp (Warsz). 2010 Oct;58(5):385-93. doi: 10.1007/s00005-010-0092-z. Epub 2010 Aug 5. PMID: 20686866.
- Reynier F, Cazalis MA, Lecoq A, Paye M, Rosa A, Durand A, Jhumka U, Mougin B, Miossec P, Bendelac N, Nicolino M, Thivolet C. Lack of association of IL-10 promoter gene variants with type 1 diabetes in a French population. Hum Immunol. 2006 Apr-May;67(4-5):311-7. doi: 10.1016/j.humimm.2006.02.029. Epub 2006 Apr 7. PMID: 16720211.
- Ide A, Kawasaki E, Abiru N, Sun F, Fukushima T, Ishii R, Takahashi R, Kuwahara H, Fujita N, Kita A, Imaizumi M, Oshima K, Usa T, Uotani S, Ejima E, Yamasaki H, Ashizawa K, Yamaguchi Y, Eguchi K. Interleukin-10 gene promoter region polymorphisms in patients with type 1 diabetes and autoimmune thyroid disease. Ann N Y Acad Sci. 2003 Nov;1005:344-7. doi: 10.1196/annals.1288.055. PMID: 14679088.
- Ide A, Kawasaki E, Abiru N, Sun F, Takahashi R, Kuwahara H, Fujita N, Kita A, Oshima K, Sakamaki H, Uotani S, Yamasaki H, Yamaguchi Y, Eguchi K. Genetic association between interleukin-10 gene promoter region polymorphisms and type 1 diabetes age-at-onset. Hum Immunol. 2002 Aug;63(8):690-5. doi: 10.1016/s0198-8859(02)00417-2. PMID: 12121678.
- Urcelay E, Santiago JL, de la Calle H, Martínez A, Figueredo A, Fernández-Arquero M, de la Concha EG. Interleukin-10 polymorphisms in Spanish type 1 diabetes patients. Genes Immun. 2004 Jun;5(4):306-9. doi: 10.1038/sj.gene.6364071. PMID: 15057267.
- Siekiera U, Jarosz-Chobot P, Janusz J, Koehler B. [Polymorphism of TNF-alpha (308 A/G), IL-10 (1082 A/G, 819 C/T 592 A/C), IL-6 (174 G/C), and IFN-gamma (874 A/T); genetically conditioned cytokine synthesis level in children with diabetes type 1]. Endokrynol Diabetol Chor Przemiany Materii Wieku Rozw 2002; 8(1): 29-34. [PMID: 12818128]
- Jahromi MM, Millward BA, Demaine AG. A polymorphism in the promoter region of the gene for interleukin-6 is associated with susceptibility to type 1 diabetes mellitus. J Interferon Cytokine Res. 2000 Oct;20(10):885-8. doi: 10.1089/10799900050163253. PMID: 11054276.
- Myśliwiec M, Myśliwska J, Zorena K, Balcerska A, Malinowska E, Wiśniewski P. Interleukin 6 -174(G>C) gene polymorphism is related to celiac disease and autoimmune thyroiditis coincidence in diabetes type 1 children. Diabetes Res Clin Pract. 2008 Oct;82(1):108-12. doi: 10.1016/j.diabres.2008.07.004. Epub 2008 Aug 9. PMID: 18692934.
- Cooper JD, Smyth DJ, Bailey R, Payne F, Downes K, Godfrey LM, Masters J, Zeitels LR, Vella A, Walker NM, Todd JA. The candidate genes TAF5L, TCF7, PDCD1, IL6 and ICAM1 cannot be excluded from having effects in type 1 diabetes. BMC Med Genet. 2007 Nov 28;8:71. doi: 10.1186/1471-2350-8-71. PMID: 18045485; PMCID: PMC2217539.
- Ishihara K, Hirano T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 2002 Aug-Oct;13(4-5):357-68. doi: 10.1016/s1359-6101(02)00027-8. PMID: 12220549.
- Kristiansen OP, Mandrup-Poulsen T. Interleukin-6 and diabetes: the good, the bad, or the indifferent? Diabetes. 2005 Dec;54 Suppl 2:S114-24. doi: 10.2337/diabetes.54.suppl_2.s114. PMID: 16306329.
- Naugler WE, Karin M. The wolf in sheep's clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends Mol Med. 2008 Mar;14(3):109-19. doi: 10.1016/j.molmed.2007.12.007. Epub 2008 Feb 7. PMID: 18261959.
- Diamond MS, Kanneganti TD. Innate immunity: the first line of defense against SARS-CoV-2. Nat Immunol. 2022 Feb;23(2):165-176. doi: 10.1038/s41590-021-01091-0. Epub 2022 Feb 1. PMID: 35105981; PMCID: PMC8935980.
- Choi SE, Choi KM, Yoon IH, Shin JY, Kim JS, Park WY, Han DJ, Kim SC, Ahn C, Kim JY, Hwang ES, Cha CY, Szot GL, Yoon KH, Park CG. IL-6 protects pancreatic islet beta cells from pro-inflammatory cytokines-induced cell death and functional impairment in vitro and in vivo. Transpl Immunol. 2004 Jun-Jul;13(1):43-53. doi: 10.1016/j.trim.2004.04.001. PMID: 15203128.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
