Journal of Gynecological Research and Obstetrics
Toward understanding the roles of matrix metallopeptidase 1 in ovarian cancer
Cassie Hobbs1, Isabella Coogan1, Jun Hee Shin1, Derek Y Yao1, Olivia Neely1, Parmida Jamshidi1, Andrew Berchuck2, Susan K Murphy1 and Zhiqing Huang1*
2Department of Obstetrics and Gynecology, Division of Gynecologic Oncology, Duke University Medical Center, Durham, NC, USA
Cite this as
Hobbs C, Coogan I, Shin JH, Yao DY, Neely O, et al. (2023) Toward understanding the roles of matrix metallopeptidase 1 in ovarian cancer. J Gynecol Res Obstet 9(1): 007-019. DOI: 10.17352/jgro.000120Copyright License
© 2023 Hobbs C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Ovarian Cancer (OC) is the leading cause of gynecologic cancer-related deaths worldwide. The leading risk factors for OC-related death are OC recurrence and the development of chemotherapy resistance. Investigation into molecular differences that distinguish primary from recurrent disease and the role of the tumor microenvironment (TME) in OC progression may help identify therapeutic targets. Gene expression microarray data comparisons between 21 primary and 21 recurrent OC specimens (16 matched pairs) showed significantly increased expression of matrix metallopeptidase 1 (MMP1) in the recurrent specimens (p = 0.03). We, therefore, investigated MMP1 expression regulation and how endogenous and exogenous MMP1 expression influences OC cell proliferation, migration/invasion and chemosensitivity. Both endogenous MMP1 knockdown and low levels of exogenous MMP1 increased cell proliferation of the OC cell line, CAOV2 (p < 0.01 and p < 0.001, respectively). Furthermore, CAOV2 cells cultured with low exogenous MMP1 exhibited increased invasion (p = 0.04 and p = 0.002, respectively, for two shRNA-conditioned mediums, shMMP1-1 or shMMP1-2) and faster migration by wound healing assay relative to controls without MMP1 knockdown. CAOV2 MMP1 knockdown cells were also more resistant than controls to carboplatin (p = 0.04) and paclitaxel (p = 0.017). To explore the functions of cancer environmental MMP1 in different cancer cells, 3 OC cell lines (CAOV2, HEYA8 and SKOV3) were tested for their proliferation when cultured under a low MMP1 conditioned medium. Interestingly, while the proliferation was increased in CAOV2 and HEYA8 cells, it was reduced when SKOV3 OC cells were cultured with low exogenous MMP1 (CAOV2: * p = 0.01, HEYA8: **** p = 0.0004, SKOV3: ** p = 0.002). These results likely reflect inherent MMP1 expression variability in OC tissues and cell lines that is at least partly dependent on other endogenous parameters of the TME, including pH, metabolic state, and oxygenation, all of which were found to alter levels of endogenous MMP1. Given the ability of MMP1 to promote oncogenic or tumor-suppressive behaviors, further study will be necessary to better understand how MMP1 contributes to promoting or restraining tumor progression in an individualized manner.
Abbreviations
MMP: matrix metallopeptidase; OC: Ovarian Cancer; TME: Tumor Microenvironment; ECM: Extracellular Matrix; ROC: Recurrent Ovarian Cancer; POC: Primary Ovarian Cancer; CM: Conditioned Medium; CTL: Control; FBS: Fetal Bovine Serum; P/S: Penicillin Streptomycin; DMEM: Dulbecco′s Modified Eagle′s Medium; RMA: Robust Multiarray Analysis; MAPK: Mitogen-Activated Protein Kinase; DAC: 5-aza-2′-Deoxycytidine; NFT: Normal Fallopian Tubes; CAF: Cancer-Associated Fibroblasts
Introduction
Ovarian Cancer (OC) is the second most common gynecologic malignancy and the fifth leading cause of cancer-related death in women in the United States [1]. Treatment strategies have not significantly improved in the past 30 years, particularly with respect to reducing disease recurrence. The main treatment for primary OC remains surgical removal of primary cancer and cytoreduction of metastases and chemotherapy using platinum-based antineoplastic drugs [2]. Neoadjuvant chemotherapy followed by interval debulking and then additional chemotherapy is also sometimes used [3]. Some patients receive maintenance treatment with poly ADP ribose polymerase (PARP) inhibitors [4]. Although most patients achieve clinical remission after surgery and chemotherapy, within a few months to several years after primary treatment, 70-90% of patients with advanced OC experience tumor recurrence, which is an incurable disease despite potentially positive initial responses to treatment [5-7]. Although the molecular alterations of primary OC have been extensively studied, little data is available on recurrent tumors, as it is difficult to obtain recurrent tumor samples, even more so matched primary-recurrent tumor pairs [7]. Investigation into the genomic differences that characterize recurrent ovarian cancer, and how these differences contribute to tumor progression and chemotherapeutic response, is of great importance in the effort to identify novel targets that will allow for more durable treatment of patients with OC.
Here we examined microarray gene expression profiles for 21 primary and 21 recurrent serous epithelial ovarian cancers, including 16 matched pairs where primary and recurrent tumor tissue samples came from the same patient. We found that the seven most differentially expressed genes between these two groups belong to the extracellular matrix (ECM) superfamily, including two matrix metallopeptidase (MMP) genes, MMP1 and MMP13. The ECM is an important contributor to the Tumor Microenvironment (TME), which has a prominent role in the development of chemoresistance [8,9]. The TME includes the blood vessels, immune cells, fibroblasts, signaling molecules, and ECM that surround a tumor [10,11]. The MMP family of proteins are zinc-dependent enzymes that function to degrade the ECM. Recent studies on MMPs have focused on their function during tissue injury in the skin and cardiovascular system [12]. MMP1 plays a role in response to DNA damage caused by radiation and DNA repair [13,14]. In response to DNA damage in the skin, soluble cytokines, including TNF-α and IL-1, stimulate dermal fibroblasts to upregulate MMP1 transcription through the p38 mitogen-activated protein kinase (MAPK) pathway [15,16]. Enhancing DNA repair can reduce MMP1 expression in human skin cells and tissues [14]. Surprisingly, given the modulation of MMP1 expression levels in association with DNA damage, no studies have yet determined if MMP1 plays a role in chemotherapeutic response [17,18]. The association between MMP1 and DNA damage is of particular importance in OC since first-line platinum-based drugs work by causing DNA damage and efficient mechanisms for DNA repair after exposure to chemotherapy are vital to the proliferation and invasion of cancer cells [19].
While there are studies investigating the role of MMP1 in ovarian cancer, no studies have reported on MMP1 in recurrent OC [20-25]. Given the very strong relationship between recurrent OC and mortality, we focused our investigation on genomic differences between primary and recurrent OC, with a long-term goal of exploiting these differences for the development of novel targeted therapies. The objective of the present study was to determine the role of endogenous and exogenous MMP1 in OC proliferation, migration/invasion and chemosensitivity and to examine how environmental characteristics of the TME regulate MMP1 expression.
Materials and Methods
Tumor Samples
We used 21 primary (POC) and 21 Recurrent OC (ROC) tissues from patients with stage III/IV serous epithelial OC from the Duke Gynecologic Oncology Tissue Bank. Of the 21 tumor pairs, 16 were matched pairs, from patients who contributed both primary and recurrent tumor samples. The primary tumor specimens were collected at the time of initial debulking surgery. Recurrent tumor tissue was obtained from the same patients during “second look” surgeries. Samples were obtained, prior to the initial surgery, after patients provided written, informed consent, in accordance with protocols approved by the Duke University Institutional Review Board.
DNA and RNA extraction
DNA and RNA were simultaneously extracted from each of the fresh-frozen tissue samples using the AllPrep DNA/RNA Mini Kit, according to the manufacturer’s protocol (Qiagen; Germantown, MD; Cat#80204). Nucleic acid concentration and purity were assessed using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific; Waltham, MA).
DNA methylation by illumina infinium humanmethylation 450 beadchip
DNA (500 ng) was bisulfite converted using the Zymo EZ DNA Methylation Kit (ZymoResearch, Tustin, CA), according to the manufacturer’s protocol. The Illumina Infinium HumanMethylation450 BeadChip was used to generate quantitative DNA methylation data using bisulfite-modified genomic DNA from the 21 POC and 21 ROC specimens. This data is publicly available through the Duke Digital Data Repository, https://doi.org/10.7924/r4765hq57 [26].
Gene expression by microarray
Affymetrix Human Genome U133A Plus 2 arrays were used by the Duke DNA Microarray Facility to quantify gene expression, using total RNA isolated from the 21 POC and 21 ROC frozen ovarian tumor samples. This data is publicly available through the Duke Digital Data Repository, https://doi.org/10.7924/r43f4sx2k [27]. Previously published Affymetrix U133A gene expression data were used from two normal fallopian tube fimbriae epithelia, 12 stages I/II serous OCs, and 55 stages III/IV serous OCs [28]. We also used our Affymetrix HT Human Genome U133A Array gene expression data (NCBI GSE25428) for 22 OC cell lines treated for 72 hours with vehicle or 5 µM 5’-aza-2’-deoxycytidine (Decitabine, DAC; Sigma-Aldrich; St. Louis, MO; #A3656).
Cell culture and treatment
Ovarian cancer cell lines CAOV2 (aka OVCAR2), HEYA8, or SKOV3 were provided by the Gynecologic Oncology Tumor Bank at Duke University Medical Center. CAOV2 was used as the main study model, as it was obtained from ascites of a patient with ovarian adenocarcinoma [29] whose tumor was inherently resistant to cisplatin [30], which is relevant to chemoresistant, recurrent ovarian cancer. Cells underwent genetic authenticity testing using the GenePrint 10 kit (Promega; Madison, WI) at the Duke University DNA Analysis Facility and testing for mycoplasma at the Duke Cell Culture Facility prior to each expansion of new frozen stocks. The cells were cultured in RPMI 1640 (Sigma Aldrich; Milwaukee, WI) supplemented with 10% fetal bovine serum (FBS) (Invitrogen; Carlsbad, CA) and 1% penicillin-streptomycin (P/S) solution (Invitrogen; Carlsbad, CA). The embryonic kidney cell line, HEK293T, was cultured with Dulbecco’s Modified Eagle’s Medium DMEM (Sigma Aldrich; Saint Louis, MO) supplemented with 10% FBS (Invitrogen; Carlsbad, CA) and 1% P/S solution (Invitrogen, Carlsbad, CA). WI38 (ATCC; Manassas, VA), a human lung fibroblast cell line, was from a normal embryo at about three months gestation. WI38 cells were cultured with EMEM (EBSS) (Sigma Aldrich, Saint Louis, MO), 2 mM glutamine (Sigma Aldrich; Saint Louis, MO), 1% non-essential amino acids (NEAA) (Invitrogen; Carlsbad, CA), 10% FBS (Invitrogen; Carlsbad, CA) and 1% P/S solution (Invitrogen; Carlsbad, CA). All cell lines were grown in a 37 °C humidified incubator with 5% atmospheric CO2. Both HEK293T and WI38 cell lines were tested for authenticity at the Duke Cell Culture Facility and were confirmed to be mycoplasma free.
Variable pH: Thirty-seven percent HCl (Millipore Sigma; St. Louis, MO) and 10N NaOH (Millipore Sigma; St. Louis, MO) solutions were added to RPMI 1640 (Sigma Aldrich; Milwaukee, WI), supplemented with 10% FBS (Invitrogen; Carlsbad, CA) and 1% P/S solution, in a dropwise fashion until the desired pH was achieved. pH was measured using a Benchtop pH-mV Meter (Cole Parmer; Vernon Hills, IL). The pH-adjusted culture medium was filtered using a 0.22 µm pore, 250 mL vacuum filter (Corning Life Sciences, Corning, NY).
Hypoxic culture: Anhydrous cobalt (II) chloride (CoCl2) (Millipore Sigma; St. Louis, MO) was used for chemical hypoxia induction. Dosing of CoCl2 was in accordance with previously published literature for establishing chemical hypoxia in breast and ovarian cancer cell lines [31]. CoCl2 was dissolved in distilled water. The resulting aqueous CoCl2 solution was directly added to the cell culture media at 100, 200, and 300 µM concentrations.
High glucose culture: D- (+)-glucose (Sigma Aldrich; Milwaukee, WI) was used to supplement glucose concentrations in the culture medium. Glucose solution was added to RPMI 1640 (Sigma Aldrich; Milwaukee, WI), supplemented with 10% FBS (Invitrogen; Carlsbad, CA) and 1% P/S solution, and stored at 4 °C for later use. Because normal blood sugar levels in vivo are approximately 5.5 mM D-glucose, we tested glucose levels in a cell culture medium ranging from 1.1 mM to as high as 33 mM for high glucose culture.
D-methionine: D-methionine (D-2-Amino-4-(methylthio) butanoic acid, (R)-2-Amino-4-(methyl mercapto) butyric acid, C5H11NO2S, Sigma Aldrich; Milwaukee, WI) was added at 0.05 mM into RPMI 1640 (Sigma Aldrich; Milwaukee, WI), supplemented with 10% FBS (Invitrogen; Carlsbad, CA) and 1% P/S solution.
MMP1 knockdown
shRNA specific to MMP1 was introduced into CAOV2, WI38 and HEK293T cells using a lentiviral transduction system according to the Addgene protocol (Watertown, MA). Briefly, HEK293T cells were transfected with 1µg of a lentivirus plasmid containing either control shRNA (Sigma Aldrich; Milwaukee, Wis., USA) or two independent MMP1-specific shRNAs (shMMP1-1: 3334, shMMP1-2: 3335; Duke Functional Genomics Shared Resource) together with packaging plasmids. FuGENE HD transfection reagent (Promega; Madison, WI) was used according to the manufacturer’s recommendations. After 48 and 72 hours of transfection, the supernatants containing the virus particles were collected and stored at -80 °C until needed. The OC cell line CAOV2, fibroblast cell line WI38 (used to create conditioned medium for MMP1 low expression), and embryonic kidney cell line HEK293T (used to create conditioned medium for MMP1 low expression), were seeded the day before infection with lentiviral shRNA into 6-well plates at a density of 1 × 105 cells/well. The cells were 60% - 70% confluent at the time of infection. The medium was replaced with medium containing 8 µg/mL of Polybrene (Sigma Aldrich; Milwaukee, WI). The virus-containing control shRNA or MMP1 shRNAs (~1 x 106 for the viral titer) from the last step was added to each cell line. After 72 hours of incubation, the cells were selected for two weeks using puromycin at 2 µg/mL for CAOV2, 1 µg/mL for WI38, and 5 µg/mL for HEK293T cells. Cells were then harvested to determine MMP1 expression using qRT-PCR analysis.
RNA extraction and qRT-PCR
Total RNA extraction from CAOV2 shCTL and CAOV2 shMMP1 knockdown cells and WI38 shCTL and WI38 shMMP1 knockdown cells was achieved using TRIzol Reagent (Invitrogen; Carlsbad, CA). RNA quantity was measured on a Nanodrop 1000 spectrophotometer (Thermo Scientific; Waltham, MA). MMP1 expression was analyzed by qRT-PCR with the one-step PCR ToughMix® (QuantaBio; Beverly, MA). Relative mRNA expression levels of MMP1 were analyzed using an MMP1 probe (hs00899658; ThermoFisher; Waltham, MA). A B2M (a housekeeping gene) probe (hs00196842; ThermoFisher; Waltham, MA) was used as an internal loading control. All assessments of MMP1 expression levels in real-time qRT-PCR experiments were performed in duplicate, normalized to housekeeping gene (B2M) levels and the data presented represent the average of those measurements.
Pyrosequencing
The pyrosequencing assay was designed to analyze the DNA methylation of the MMP1 gene at its promotor region. Four CGs at the MMP1 promotor region were included in the sequence to analyze, 5’-GAA TTT YGA AGA GTT ATY GTA AAG TGA GTG TTG GGG GAG TTG AAT TTT AGT TAG TAT AGG TGT YGA ATA GTT ATT AGG TGY GTA GTG TTA GTA ATT TTA TTT TTT GTT T-3’. PCR was carried out with bisulfite-modified genomic DNA with primers, F: 5’-AGG TAG TTT AAT AAA GGT AGA AGG G-3’, and biotinylated R primer: 5’-btn-AAT TTC TCC ACA CAC CTT ACT C-3’ (Sigma Aldrich; Milwaukee, WI). Defined mixtures of fully methylated and unmethylated human genomic DNAs (EpiTect Control DNA; Qiagen) were used to validate the performance of the pyrosequencing assay. Bisulfite conversion was performed using 800 ng genomic DNA from OC cells using Zymo EZ DNA Methylation Kit (ZymoResearch, Tustin, CA). Bisulfite-converted DNAs (20 ng) were used in a total PCR reaction volume of 20 µL. PCR was performed at 95 ºC for 15 minutes followed by 55 cycles of 95 ºC for 30 seconds/65 ºC for 30 seconds/72 ºC for 30 seconds, with a final extension at 72 ºC for 10 minutes. The pyrosequencing was performed with the sequencing primer, 5’-GGT AGA AGG GAA TTT TAG A-3’, with a PyroMark Q96 MD Pyrosequencer (Qiagen, CA).
Generation of conditioned medium
HEK293T and WI38 cells transfected with MMP1-specific shRNAs or control shRNA were seeded in 100-mm cell culture dishes at a density of 2.0 x 106 cells/plate with DMEM (Sigma Aldrich; Saint Louis, MO) supplemented with 10% FBS (Invitrogen; Carlsbad, CA) and 1% P/S solution (Sigma Aldrich; Saint Louis, MO). Cells were incubated at 37 °C in a humidified atmosphere with 5% CO2. Twenty-four hours later, the medium was collected and centrifuged to remove cells and cell debris. The supernatant was collected and used as the MMP1 knockdown conditioned medium, labeled shMMP1 CM for the conditioned medium from HEK293T or WI38 cells. The non-silencing control medium obtained from shCTL-transfected HEK293T or WI38 cells was labeled as shCTL CM for the control conditioned medium from HEK293T or WI38 cells.
ELISA assay
HEK293T cells transfected with MMP1-specific shRNAs (shMMP1-1) or control shRNA (shCTL) were seeded in 100-mm cell culture dishes at a density of 2.0 x 106 cells/plate with DMEM (Sigma Aldrich; Saint Louis, MO) supplemented with 10% FBS (Invitrogen; Carlsbad, CA) and 1% P/S solution (Sigma Aldrich; Saint Louis, MO). Cells were incubated at 37 °C in a humidified atmosphere with 5% CO2. Twenty-four hours later, the medium was collected and centrifuged to remove cells and cell debris. The medium was used for measuring MMP-1 by ELISA assay, using the kit from Invitrogen (human MMP-1, Cat# EHMMP1) in accordance with the manufacturer’s instructions. The protein concentration was determined using Bradford protein assay, according to the instructions from the manufacturer (Bio-Rad). The test was done in triplicate for each CM from shMMP1-1 or shCTL.
Cell proliferation assay with MMP1 knockdown cells and MMP1 low-conditioned medium
CAOV2 cells transfected with either control shRNA (shCTL) or MMP1-specific shRNA were cultured in the clear bottom, black 96-well plates (Corning Life Science; Corning, NY) in RPMI-1640 medium, with 10% FBS and 1% P/S, at 5,000 cells/well. After 48 hours, 50 µL of CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega Corporation; Madison, WI) was added to each well. The luminescence signal was measured using a microplate reader. Experiments were performed twice with three replicates for each test.
CAOV2 cells were cultured in 96-well plates with conditioned medium (CM) from MMP1 knockdown WI38 cells, shMMP1 CM, or an off-target control shRNA, shCTL CM. After 48 hours, 50 µL of CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega Corporation; Madison, WI) was added to each well. The luminescence signal was measured using a microplate reader. Experiments were performed twice with four replicates for each test.
Chemosensitivity test with MMP1 low-conditioned medium
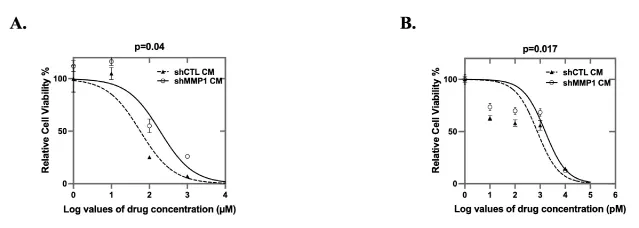
CAOV2 cells were cultured in the clear bottom, black 96-well plates (Corning Life Science; Corning, NY) in RPMI-1640 medium with 100 µL conditioned medium (CM) collected from WI38 cells transfected with shCTL or shMMP1. After 24 hours, the medium was replaced with a fresh conditioned medium containing carboplatin or paclitaxel. The concentration range for paclitaxel was 1 nM to 10 µM in 10-fold increments. For carboplatin, the range was 1 µM to 1000 µM in 10-fold increments. Both drugs were diluted in RPMI-1640 medium with 10% FBS and 1% P/S in a total volume of 100 µL per well. After 48 hours, 50 µL of CellTiter-Glo® Luminescent Cell Viability Assay (Promega Corporation; Madison, WI) was added to each well. The luminescence signal was measured using a microplate reader. Experiments were performed twice with three replicates for each test.
Wound healing assay
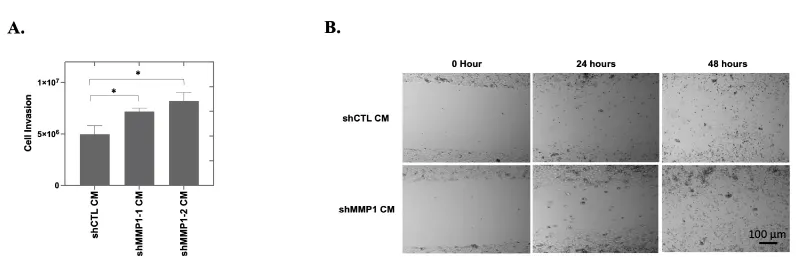
For the wound healing experiments with the conditioned medium, we seeded equal numbers of CAOV2 cells into two wells of 6-well plates at a density of 3 x 105 cells/well using the conditioned medium from WI38 shCTL or WI38 shMMP1 (shCTL CM or shMMP1 CM). After 24 hours, the cells reached 90% - 95% confluence. A uniform “wound” was then generated by scratching a sterile 1 mL pipet tip across each cell monolayer. Micrographs for wounds and wound healing were taken to capture the process of cell migration starting at the time the scratch was introduced (time 0) until closure. Micrographs for wounds and wound healing were taken to capture the process of cell migration at times 0, 24 and 48 hours after “wounding”. These experiments were independently performed twice.
Cell invasion assay
Invasion potential for CAOV2 cells was assessed using a 96-well Invasion Assay (Cell Biolabs; San Diego, CA). CAOV2 cells were seeded at 5 x 104 cells/well in the top chamber of the assay dish with WI38 shCTL CM, shMMP1-1 CM, or shMMP1-2 CM. The bottom chamber of the assay dish contained either conditioned medium collected from shMMP1-1, shMMP1-2, or control (shCTL) WI38 cells containing 20% FBS. Each condition was performed in triplicate. The plate was incubated at 37°C in a 5% CO2 humidified incubator for 48 hours. The cells in the bottom chamber were stained using the reagents provided in the kit and the absorbance at 560 nm was measured using a microplate reader.
Statistical analysis
The microarray data from primary and recurrent OC samples were adjusted for the p values for multiple gene expression comparisons. The microarray data from mock-treatment or treatment of OC cell lines with 5-aza-2′-deoxycytidine (DAC) was analyzed using Student’s t-test with GraphPad Prism 8 to compare MMP1 gene expression. The microarray gene expression data from early OC, advanced OC and normal fallopian tube (NFT) samples were compared for MMP1 expression using a Student t-test. The microarray data, from primary and recurrent OC samples, were analyzed by the Wilcoxon test to compare the gene expression between 21 unpaired primary and recurrent OC samples and between 16 matched primary and recurrent OC pairs. Heatmapper (http://www.heatmapper.ca) was used for the generation of heatmaps [32]. Results from cell proliferation, invasion and qRT-PCR assays are expressed as means +/- standard deviation (SD). Student’s t-test was used to assess two group differences for continuous variables. The cell viability assays for cells cultured with shCTL CM or shMMP1 CM and treated with chemotherapeutic drugs were analyzed and compared between groups using a One-Way ANOVA assay. For all data analyses, a p - value <0.05 was considered statistically significant.
Results
MMP1 expression in fallopian tube precursor cells versus primary serous OC
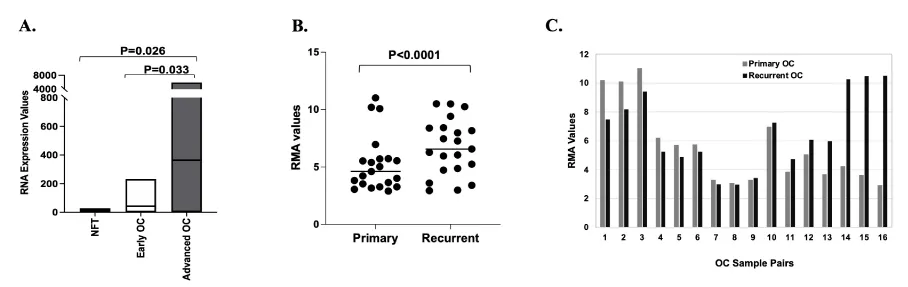
Previous studies have indicated that MMP1 expression is upregulated in OC as compared to normal ovarian tissue specimens [20]. However, abundant data now support that the site of origin for serous epithelial OC is the fallopian tube epithelia rather than ovarian surface epithelial cells [33,34]. To better understand the level of MMP1 expression in primary OC as compared to the non-diseased tissue of origin, we analyzed existing Affymetrix U133A microarray data from 67 serous OCs, including 55 stages III/IV cases (30 who lived <3 years and 25 who lived >7 years) and 12 stages I/II cases [28]. These results were compared to microarray data from the fallopian tube epithelia of two patients without the disease. Advanced OC had significantly higher expression of MMP1 compared to the normal fallopian tube (Figure 1A, p = 0.026), which is consistent with findings from other researchers (19). MMP1 also showed significantly higher expression in advanced OC compared to early-stage OC (Figure 1A, p = 0.033). We found that the minimal expression in early-stage tumors was Robust Multiarray Analysis (RMA) = 0.04 and maximal expression was RMA = 2.37, a range of 2.32, whereas the minimal MMP1 expression in advanced tumors was RMA = 0.11 and maximal expression was RMA = 3.77, range of 3.66. The data suggested that the early-stage OCs have narrower MMP1 expression ranges than do the advanced tumors.
Differentially expressed genes in primary versus recurrent ovarian cancer
Analysis of gene expression microarray data from 21 primary (POC) and 21 recurrent ovarian cancer (ROC) samples identified about 900 genes showing significant differences in expression between POC and ROC (unadjusted p < 0.05). Seven of these showed >two-fold higher expression in ROC and five showed >two-fold higher expression in POC (data not shown). Interestingly, all seven of the upregulated genes in ROC belong to the ECM superfamily, including Periostin (POSTN), Collagen 11A1 (COL11A1), Tenascin C (TNC), Asporin (ASPN), Epiphycan (EPYC), matrix metallopeptidase 13 (MMP13), and matrix metallopeptidase 1 (MMP1). These results support the importance of the ECM, a component of the TME, in ROC. The MMP1 expression differences between the 21 POC and 21 ROC are shown in Figure 1B (p < 0.0001). Among the 21 POC and 21 ROC samples, there were 16 primary-recurrent paired sets each obtained from the same patient (Figure 1C). Although changes in MMP1 expression were observed across the two distinct disease time points for each pair, the direction of change was not consistent for each pair of POC and ROC (Figure 1C), with 8 pairs showing decreased and 8 pairs showing increased expression of MMP1 in the ROCs (primary OC mean = 5.547, recurrent OC mean = 6.562, p < 0.0001, Wilcoxon test). This seemingly conflicting data suggest that the importance of MMP1 expression and thereby its functional role, depends on the individual tumor and that this logically would contribute to the large heterogeneity in phenotypes, tumor aggressiveness, and the therapeutic response observed in different individuals.
MMP1 expression is regulated by DNA methylation in ovarian cancer cells
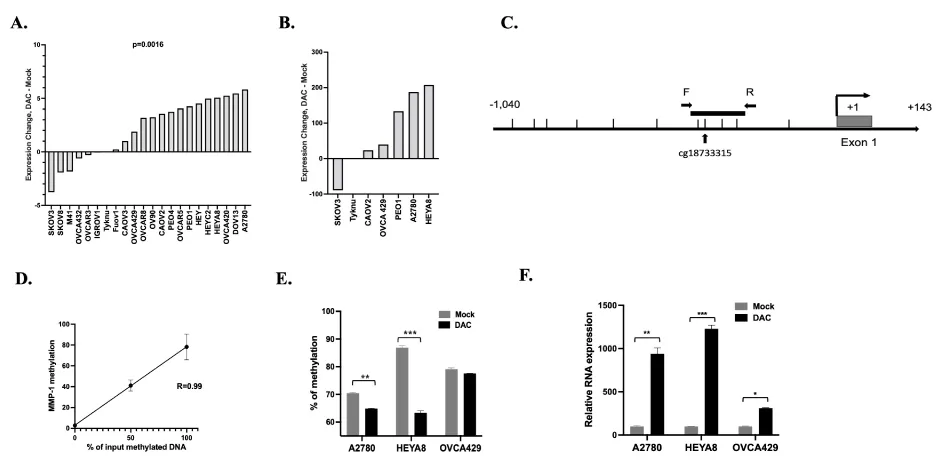
Differences in the epigenetic profiles at MMP1 could mechanistically contribute to the wide variability in MMP1 expression observed in the OC tissues. Epigenetic changes are important contributors to cancer phenotype through altered regulation of cancer-related genes [35]. We, therefore, investigated whether MMP1 gene expression is regulated by DNA methylation. Twenty-two OC cell lines were treated with 5 µM decitabine (DAC), a DNA methyltransferase inhibitor that has been used clinically for many types of cancer including OC [36]. RNA extraction was performed after 72 hours of mock treatment or treatment with DAC, followed by the generation of gene expression microarray data using the Affymetrix HT Human Genome U133A Array. Following DAC treatment, 15 of the 22 tested cell lines (68%) showed an increase in MMP1 expression, indicating that MMP1 is regulated directly or indirectly by DNA methylation in these cells (p = 0.0016, Figure 2A). To validate the microarray data, we performed qRT-PCR (Figure 2B) with seven OC cell lines that were mock-treated or treated with 5 µM DAC for 72 hours. We were able to validate the gene expression microarray findings for 6 out of 7 tested cell lines, with upregulation of MMP1 expression in the same seven showing upregulation by microarrays (Figure 2B).
To further confirm the relationship between DNA methylation and gene expression for MMP1, we designed a pyrosequencing assay to analyze four CpG sites at the MMP1 promoter region, which includes one CG (cg18733315) which is also included on the Illumina Infinium HumanMethylation 450 BeadChip as shown in Figure 2C, the schematic graph of the MMP1 promoter region. This region for pyrosequencing assay is about 400 bp upstream of the MMP1 transcription start site. Pyrosequencing assay validation was performed using fully methylated and unmethylated genomic DNAs, including 0%, 50% and 100%. The average percent methylation of the four CGs were used for analysis. There was a high correlation between the input methylated DNA and measured levels of DNA methylation (R = 0.99) supporting the ability of the pyrosequencing assay to quantify methylation levels across the possible linear range (Figure 2C). Pyrosequencing of the MMP1 promoter and qRT-PCR was performed in three cell lines: A2780, HEYA8 and OVCA429, which showed higher MMP1 DNA methylation in mock-treated cells which was reduced with DAC treatment (Figure 2D) and a concomitant significant increase in MMP1 expression (A2780 p < 0.001, HEYA8 p < 0.01 and OVCA429 p < 0.05, Figure 2E). Our data support that the expression of MMP1 is at least partially regulated directly by DNA methylation.
MMP1 expression in cancer cells is affected by the TME
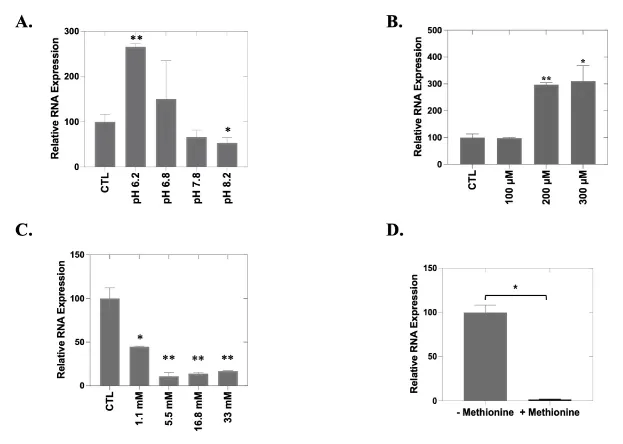
Soluble factors secreted by the TME can contribute to abnormal proliferation, angiogenesis, metastasis and drug resistance [37,38]. Previous studies have demonstrated that cultured cancer cells upregulate the secretion of MMPs in the presence of conditioned media from tumor cells [39]. To determine how MMP1 expression is influenced by the local microenvironment, we performed a series of experiments in which we altered pH, induced hypoxia, or provided additional glucose or methionine. We found that expression levels of MMP1 indeed vary with the pH of the culture medium. Significant increased MMP1 expression was observed under more acidic conditions (pH 6.2; p = 0.003) while MMP1 repression was observed under more basic conditions (pH 8.2; p = 0.04) (Figure 3A). Hypoxic conditions were induced by increasing the CoCl2 concentration in the cell culture medium, which tripled expression levels of MMP1 (200 µM, p = 0.002; 300 µM, p = 0.02) (Figure 3B). We also observed a dose-dependent reduction in MMP1 expression with increasing glucose concentration (p < 0.05; Figure 3C). Cells cultured in a methionine-depleted medium showed a nearly two-log increase in MMP1 expression as compared with cells cultured with supplemental methionine (p = 0.036; Figure 3D). These data support that the level of MMP1 expression in OC cells is responsive to, and greatly influenced by, specific and relevant TME cues.
MMP1 repression in cancer cells and the TME enhances proliferation and migration
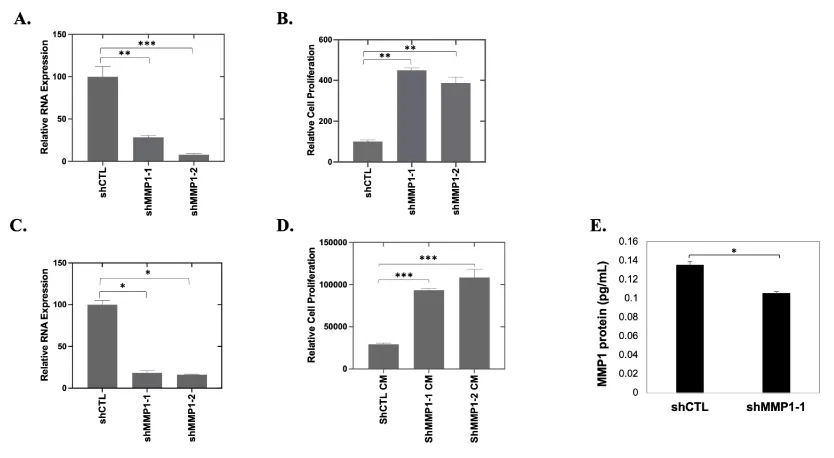
To determine the effect of reducing the availability of endogenous MMP1 in OC cancer cells, knockdown of MMP1 expression in CAOV2 cells (Figure 4A) followed by proliferation analysis (Figure 4B) was performed. Contrary to existing data from other researchers using other cancer cell lines [21,22,40], CAOV2 showed significantly increased cell proliferation (Figure 4B, p < 0.01) when MMP1 was downregulated in cancer cells using two shRNAs specific to MMP1 (shMMP1-1 and shMMP1-2) or an off-target control shRNA (shCTL) (Figure 4A, p < 0.01 for shMMP1-1 and p < 0.001 for shMMP1-2). To further examine the functions of exogenous MMP1, like that produced in the ECM, we prepared conditioned media having low MMP1 from WI38 cells in which MMP1 was targeted for knockdown with shRNA (shMMP1 CM). QRT-PCR data confirmed the efficacy of shRNA-mediated knockdown of MMP1 in WI38 cells using two shRNAs specific to MMP1 (shMMP1-1 and shMMP1-2) or an off-target control shRNA (shCTL). Knockdown efficiency was ~80% MMP1 as determined by qRT-PCR (*p = 0.04 and * p = 0.03 for shMMP1-1 and shMMP1-2, respectively; Figure 4C) when compared with the WI38 cells transfected with off-target control shRNA. CAOV2 cells cultured with the shMMP1 CM showed increased proliferation relative to cells in the shRNA control CM (shCTL CM), which had relatively higher MMP1 expression (p < 0.001 for both shMMP1-1 CM and shMMP1-2 CM; Figure 4D). The MMP1 protein expression in shMMP1 conditioned medium (CM) obtained from MMP1 knockdown HEK293T cells was tested using ELISA assay (p = 0.0122) (Figure 4E).
Higher expression of MMPs is associated with increased invasion in many types of cancer [40,41]. Increased MMP secretion (such as MMP2 in breast cancer) may assist in the breakdown of the basement membrane and lead to enhanced cancer cell invasion [42]. To determine the potential involvement of MMP1 in OC cell invasion, we performed invasion assays using CAOV2 cells cultured with WI38 shMMP1 or shCTL CM. Knockdown of MMP1 expression was confirmed in WI38 cells using qRT-PCR (Figure 4C). CAOV2 cells exhibited a more invasive phenotype when cultured with shMMP1 CM (p = 0.04 and p = 0.002, respectively for shMMP1-1 or shMMP1-2 conditioned medium; Figure 5A). Similar results were achieved with CM from HEK293T cell cells (Data not shown).
MMP1 is reported to promote cancer metastasis when secreted into the ECM by cancer cells [12,21,22]. To assess the effect of reducing exogenous MMP1 on OC cell migration, we seeded 3 x 105 CAOV2 cells/well and added MMP1 low CM (shMMP1 CM) or MMP1 control (shCTL CM) from WI38 cells (refer to Figure 4C for knockdown efficiency). After 24 hours of culture, we introduced a gap in each cell monolayer using a sterile pipet tip and documented cell migration to close the gap with micrographs from time 0 to gap closure at 48 hours. Like the MMP1 knockdown cells, CAOV2 cells cultured with shMMP1 CM were able to close the gap more rapidly as compared to cells cultured with shCTL CM (Figure 5B). Together and in agreement with the cell proliferation data, we found that low endogenous and exogenous MMP1 levels are associated with increased invasion and migration.
MMP1 repression in OC cells enhances OC cell chemoresistance
TME factors have emerged as suggested key players in the development of chemoresistance and malignant progression [43]. We performed cell viability assays (MTS) after chemotherapeutic drug treatment for CAOV2 cells cultured under MMP1 low-conditioned medium (shMMP1 CM) or shRNA control conditioned medium (shCTL CM) (Figure 6). The MMP1 conditioned medium was generated from WI38 cells transduced by shCTL or shMMP1 (refer to Figure 4C for knockdown efficiency) and the cells were further treated with (Figure 6A) carboplatin (from 1 µM to 1000 µM in 10-fold increments) or (Figure 6B) paclitaxel (from 1 nM to 10 µM in 10-fold increments). The IC50 values for paclitaxel treatment were 768.2 nM for CAOV2 in shCTL CM and 1.587µM in shMMP1 CM. The IC50 values for carboplatin treatment were 57.49 µM for CAOV2 in shCTL CM and 190.8µM in shMMP1 CM. Consistent with the cell proliferation and invasion data, CAOV2 under shMMP1 CM showed significantly more resistance to chemotherapy treatment (p = 0.04 and p = 0.017 for carboplatin and paclitaxel treatment, respectively) when each was compared to cell viability for CAOV2 cultured under shCTL CM.
Differential effects of MMP1 on OC cell proliferation
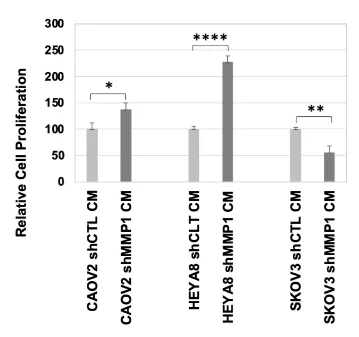
The finding that low MMP1 expression significantly increased cancer cell proliferation and migration was seemingly contrary to the findings from OC clinical samples. To determine if the in vitro results were cell line dependent, we tested the proliferation of CAOV2, HEYA8 and SKOV3 OC cells cultured under shMMP1 or shCTL CM from WI38 cells (Figure 7). The results showed cell proliferation was significantly increased in CAOV2 and HEYA8 cells (p = 0.01 and p = 0.0004 for CAOV2 and HEYA8, respectively), but reduced proliferation was seen in SKOV3 cells when they were cultured in low MMP1 conditions (p = 0.002) (Figure 7).
Discussion
This study examined the role of the TME in the regulation of MMP1 expression in OC. MMP1 expression was found to be higher in ROC and in tumors from OC patients who had short-term versus long-term survival post-diagnosis. We also showed that MMP1 expression in OC cells is regulated epigenetically by DNA methylation at the promoter region. We showed that the expression of MMP1 is significantly altered by changes relevant to the conditions in the TME, including pH, oxygen concentration and nutrient status in the culture media. Somewhat unexpectedly, we found that decreased availability of exogenous MMP1 leads to a more aggressive cancer cell phenotype, characterized by increased cell proliferation, increased invasion/migration, and increased resistance to chemotherapeutic agents. Taken together, our data suggest that MMP1 has different roles in ovarian cancer that are exquisitely sensitive to conditions within the TME and likely depend on the anatomical context of the cancer cells (e.g., in the tumor versus in the ascites) which warrant further studies.
Epigenetic modifications and their role in tumorigenesis are active areas of research. Using cancer-specific epigenetic changes as biomarkers for the detection and targeting of epigenetic readers and writers is a promising strategy used in cancer therapy [44,45]. In this study, we demonstrated increased MMP1 expression in most OC cell lines in response to treatment with the DNA methyltransferase inhibitor, DAC, indicating that MMP1 itself, and/or an upstream regulator(s) of MMP1, is repressed by DNA methylation. We then showed that DAC treatment is accompanied by a decrease in methylation at the MMP1 promoter along with increased expression, indicating a direct functional role for methylation at the MMP1 promoter. The DAC-mediated upregulation of MMP1 expression in OC cells combined with our results shows that low levels of MMP1 in OC are associated with a more aggressive phenotype, consistent with the idea that MMP1 plays a tumor-suppressive role in serous epithelial ovarian cancer.
Matrix metalloproteinases (MMPs) can be secreted by both cancer cells and cancer-associated fibroblasts (CAFs), suggesting that the actions of cancer cells and the TME can be synergistic in regulating cancer progression [46]. Studies have suggested that MMP factors have dual roles in cancer progression, either promoting cancer growth and invasion by degrading the cancer matrix and enhancing angiogenesis [47] or, conversely, inhibiting cancer growth by limiting tumor neovascularization [48]. We found higher MMP1 expression in ROC and in tumors from advanced OC patients, which is consistent with a more oncogenic role of MMP1. However, we also found that low exogenous MMP1 was associated with increased CAOV2 cell proliferation, cell invasion and cell migration. Although this is consistent with findings examining MMP1 knockdown in dermal fibroblasts [49], other studies have reported a more proliferative cancer phenotype when MMPs are overexpressed. This may be explained through a mechanism of releasing and activating growth factors, such as insulin-like growth factors (IGFs) and vascular endothelial growth factors (VEGF) [50,51]. MMPs are well-known key factors involved in ECM degradation that induce initiation in both physiological and pathological processes of angiogenesis [52]. However, the experimental evidence thus far demonstrates that MMPs also play a decisive role in the activation of pro-angiogenic and, in some cases, anti-angiogenic factors in cancer tissues [53]. Thus, MMPs can be considered angio-modulators, which could control new vessel formation necessary for cancer growth, progression, and spread. Therefore, MMPs have been speculated to participate in cancer angiogenesis in a cell context-dependent manner. Indeed, we found that low exogenous MMP1 enhanced proliferation for CAOV2 and HEYA8 cells but reduced proliferation for SKOV3 cells (Figure 7). The CAOV2 cell line was derived from a metastatic site ascites fluid of a patient with an aggressive OC (NCIt: C4908)[54] and HEYA8 cells were derived from high-grade ovarian serous adenocarcinoma (NCIt: C105555) [54]. SKOV3 is a cell line with epithelial morphology that was isolated from the ovary of a 64-year-old, white female with ovarian adenocarcinoma (ATCC, HTB-77). This cell line is moderately well-differentiated adenocarcinoma consistent with ovarian primary cells. Differences in the cellular response to the low MMP1 CM could also be a function of the overall available levels of endogenous plus exogenous MMP1. In this regard, it is noteworthy that MMP1 transcription was enhanced in both CAOV2 and HEYA8 cells by treatment with decitabine but repressed in SKOV3 cells (Figure 2A and 2B). This supports the idea that CAOV2 and HEYA8 cells may have been inherently more responsive to differences in exogenous MMP1 levels if endogenous levels were at least partially repressed by DNA methylation. In the context of low relative levels of endogenous MMP1, exogenous MMP1 appears to play a role in suppressing aggressive cell behaviors. Higher endogenous levels of MMP1 like that in SKOV3 cells may render reduction of exogenous MMP1 irrelevant such that the normal repressive function is still evident. The seeming contradiction between our in vitro and in vivo findings might be explained by the enriched cellular content of the tumor tissues analyzed, which does not provide meaningful information about exogenous MMP1. Our results therefore altogether support the idea that exogenous MMP1 normally helps suppress oncogenic behaviors of ovarian cancer cells but that higher endogenous MMP1 levels may override this effect. It is known that tumor cells have extensive heterogeneity in their metabolism and phenotype relative to normal tissue across cancer types. Abnormalities coming from the tumor cells are tissue-specific, leading to adaptation to the microenvironment where they developed. The interesting observation of the MMP family is the large, robust specificity profile, which suggests that its role is controlled in a tissue-specific manner; that is, MMP types are expressed according to the regulatory proteins needed for the tissue.
A prior report showed that inhibition of MMP2 and MMP9 followed by cisplatin treatment resulted in a more cytotoxic effect in OC than cisplatin alone [55]; however, there are no published studies examining the impact of MMP1 on chemotherapy resistance in OC. Our data, indicating that low levels of exogenous MMP1 result in increased chemoresistance in OC is contrary to what has been suggested for other MMP factors, but is consistent with the summation of other phenotypes shown in this paper: reduced expression of exogenous MMP1 was associated with more proliferative and invasive ovarian cancer cells. Additionally, we found MMP1 expression was associated with cell environmental changes, such as local pH, hypoxia status, glucose level and methionine level alterations. Thus, regulating the local pH, hypoxia, or restricting methionine could be useful for anti-cancer treatment by exploiting their effects on MMP1 expression and function(s).
The tumor immune microenvironment is distinct between primary and recurrent OC [56]. Immunologic factors in the TME evolve, such that factors associated with a poor prognosis in primary tumors can be associated with a positive prognosis in recurrent tumors [12]. For example, higher numbers of regulatory T cells (Tregs) in POC are associated with a decreased time to recurrence; yet higher levels of Tregs in recurrent tumors are correlated with longer overall survival [56]. Given these results, expanded studies are needed to explore potential functional differences in MMP1 in primary and recurrent OCs and the relationship between endogenous and exogenous MMP1 levels in vivo. Our results demonstrate that MMP1 has important roles in OC and provide justification for deeper exploration into mechanisms that could lead to novel therapeutic approaches.
We thank the Duke Flow Cytometry Facility and Duke Cell Culture Facility for their assistance with this work.
Consent for publication
All authors consent to publication.
Author contributions
CH generated data, performed statistical analyses and drafted the manuscript. ZH designed and supervised the study and helped draft the manuscript. SKM, IC and AB reviewed and edited the manuscript. DY generated data and performed statistical analyses. ON, CG and PJ generated data. All authors had access to the study data, contributed to data interpretation, critically reviewed the manuscript and reviewed and approved the final manuscript.
Funding statement
This study was supported by funding to ZH from the Department of Obstetrics and Gynecology at Duke University School of Medicine and the Gail Parkins Memorial Ovarian Cancer Awareness Fund. The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
- Society AC. Key Statistics for Ovarian Cancer. https://www.cancer.org/cancer/ovarian-cancer/about/key-statistics.html. 2020.
- Marchetti C, Muzii L, Romito A, Benedetti Panici P. First-line treatment of women with advanced ovarian cancer: focus on bevacizumab. Onco Targets Ther. 2019 Feb 8;12:1095-1103. doi: 10.2147/OTT.S155425. PMID: 30799939; PMCID: PMC6371937.
- Wang, D., et al., Choosing the right timing for interval debulking surgery and perioperative chemotherapy may improve the prognosis of advanced epithelial ovarian cancer: a retrospective study. Journal of Ovarian Research, 2021. 14(1): p. 49.
- Lea J. PARP-1 inhibitors can reduce ovarian cancer recurrence risk by 70% in half of patients. Cancer; Women's Health. 2021.
- Chatterjee M, Hurley LC, Levin NK, Stack M, Tainsky MA. Utility of paraneoplastic antigens as biomarkers for surveillance and prediction of recurrence in ovarian cancer. Cancer Biomark. 2017 Dec 6;20(4):369-387. doi: 10.3233/CBM-170652. PMID: 29125478; PMCID: PMC6033268.
- Morgan RJ Jr, Armstrong DK, Alvarez RD, Bakkum-Gamez JN, Behbakht K, Chen LM, Copeland L, Crispens MA, DeRosa M, Dorigo O, Gershenson DM, Gray HJ, Hakam A, Havrilesky LJ, Johnston C, Lele S, Martin L, Matulonis UA, O'Malley DM, Penson RT, Percac-Lima S, Pineda M, Plaxe SC, Powell MA, Ratner E, Remmenga SW, Rose PG, Sabbatini P, Santoso JT, Werner TL, Burns J, Hughes M. Ovarian Cancer, Version 1.2016, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2016 Sep;14(9):1134-63. doi: 10.6004/jnccn.2016.0122. PMID: 27587625.
- Christie EL, Bowtell DDL. Acquired chemotherapy resistance in ovarian cancer. Ann Oncol. 2017 Nov 1;28(suppl_8):viii13-viii15. doi: 10.1093/annonc/mdx446. PMID: 29232469.
- Senthebane DA, Jonker T, Rowe A, Thomford NE, Munro D, Dandara C, Wonkam A, Govender D, Calder B, Soares NC, Blackburn JM, Parker MI, Dzobo K. The Role of Tumor Microenvironment in Chemoresistance: 3D Extracellular Matrices as Accomplices. Int J Mol Sci. 2018 Sep 20;19(10):2861. doi: 10.3390/ijms19102861. PMID: 30241395; PMCID: PMC6213202.
- Velaei K, Samadi N, Barazvan B, Soleimani Rad J. Tumor microenvironment-mediated chemoresistance in breast cancer. Breast. 2016 Dec;30:92-100. doi: 10.1016/j.breast.2016.09.002. Epub 2016 Sep 23. PMID: 27668856.
- Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015 Apr 3;348(6230):74-80. doi: 10.1126/science.aaa6204. PMID: 25838376.
- Spill F, Reynolds DS, Kamm RD, Zaman MH. Impact of the physical microenvironment on tumor progression and metastasis. Curr Opin Biotechnol. 2016 Aug;40:41-48. doi: 10.1016/j.copbio.2016.02.007. Epub 2016 Mar 2. PMID: 26938687; PMCID: PMC4975620.
- Agarwal A, Tressel SL, Kaimal R, Balla M, Lam FH, Covic L, Kuliopulos A. Identification of a metalloprotease-chemokine signaling system in the ovarian cancer microenvironment: implications for antiangiogenic therapy. Cancer Res. 2010 Jul 15;70(14):5880-90. doi: 10.1158/0008-5472.CAN-09-4341. Epub 2010 Jun 22. PMID: 20570895; PMCID: PMC2917243.
- Kimura-Ohba S, Yang Y. Oxidative DNA Damage Mediated by Intranuclear MMP Activity Is Associated with Neuronal Apoptosis in Ischemic Stroke. Oxid Med Cell Longev. 2016;2016:6927328. doi: 10.1155/2016/6927328. Epub 2016 Jan 27. PMID: 26925194; PMCID: PMC4748094.
- Dong KK, Damaghi N, Picart SD, Markova NG, Obayashi K, Okano Y, Masaki H, Grether-Beck S, Krutmann J, Smiles KA, Yarosh DB. UV-induced DNA damage initiates release of MMP-1 in human skin. Exp Dermatol. 2008 Dec;17(12):1037-44. doi: 10.1111/j.1600-0625.2008.00747.x. Epub 2008 May 3. PMID: 18459971.
- Lindner D, Zietsch C, Becher PM, Schulze K, Schultheiss HP, Tschöpe C, Westermann D. Differential expression of matrix metalloproteases in human fibroblasts with different origins. Biochem Res Int. 2012;2012:875742. doi: 10.1155/2012/875742. Epub 2012 Mar 4. PMID: 22500233; PMCID: PMC3303709.
- Kim HL, Woo SM, Choi WR, Kim HS, Yi C, Kim KH, Cheng J, Yang SH, Suh JW. Scopoletin downregulates MMP‑1 expression in human fibroblasts via inhibition of p38 phosphorylation. Int J Mol Med. 2018 Oct;42(4):2285-2293. doi: 10.3892/ijmm.2018.3757. Epub 2018 Jul 4. PMID: 30015831.
- Nishikawa A, Iwasaki M, Akutagawa N, Manase K, Yamashita S, Endo T, Kudo R. Expression of various matrix proteases and Ets family transcriptional factors in ovarian cancer cell lines: correlation to invasive potential. Gynecol Oncol. 2000 Nov;79(2):256-63. doi: 10.1006/gyno.2000.5944. PMID: 11063654.
- Behrens P, Rothe M, Florin A, Wellmann A, Wernert N. Invasive properties of serous human epithelial ovarian tumors are related to Ets-1, MMP-1 and MMP-9 expression. Int J Mol Med. 2001 Aug;8(2):149-54. doi: 10.3892/ijmm.8.2.149. PMID: 11445865.
- Caccuri F, Sommariva M, Marsico S, Giordano F, Zani A, Giacomini A, Fraefel C, Balsari A, Caruso A. Inhibition of DNA Repair Mechanisms and Induction of Apoptosis in Triple Negative Breast Cancer Cells Expressing the Human Herpesvirus 6 U94. Cancers (Basel). 2019 Jul 18;11(7):1006. doi: 10.3390/cancers11071006. PMID: 31323788; PMCID: PMC6679437.
- Wang FQ, Fisher J, Fishman DA. MMP-1-PAR1 axis mediates LPA-induced epithelial ovarian cancer (EOC) invasion. Gynecol Oncol. 2011 Feb;120(2):247-55. doi: 10.1016/j.ygyno.2010.10.032. Epub 2010 Nov 20. PMID: 21093894.
- Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa M, Ikeda SI, Kato T, Kiyono T, Takeshita F, Kajiyama H, Kikkawa F, Ochiya T. Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat Commun. 2017 Mar 6;8:14470. doi: 10.1038/ncomms14470. PMID: 28262727; PMCID: PMC5343481.
- Yu J, Xu Z, Guo J, Yang K, Zheng J, Sun X. Tumor-associated macrophages (TAMs) depend on MMP1 for their cancer-promoting role. Cell Death Discov. 2021 Nov 9;7(1):343. doi: 10.1038/s41420-021-00730-7. PMID: 34753916; PMCID: PMC8578434.
- Cabral-Pacheco GA, Garza-Veloz I, Castruita-De la Rosa C, Ramirez-Acuña JM, Perez-Romero BA, Guerrero-Rodriguez JF, Martinez-Avila N, Martinez-Fierro ML. The Roles of Matrix Metalloproteinases and Their Inhibitors in Human Diseases. Int J Mol Sci. 2020 Dec 20;21(24):9739. doi: 10.3390/ijms21249739. PMID: 33419373; PMCID: PMC7767220.
- Seyed Hosseini E, Alizadeh Zarei M, Tarrahimofrad H, Zamani J, Haddad Kashani H, Ahmad E, Nikzad H. Synergistic effects of dendrosomal nanocurcumin and oxaliplatin on oncogenic properties of ovarian cancer cell lines by down-expression of MMPs. Biol Res. 2023 Jan 20;56(1):3. doi: 10.1186/s40659-023-00412-x. PMID: 36658640; PMCID: PMC9854214.
- Escalona RM, Chu S, Kadife E, Kelly JK, Kannourakis G, Findlay JK, Ahmed N. Knock down of TIMP-2 by siRNA and CRISPR/Cas9 mediates diverse cellular reprogramming of metastasis and chemosensitivity in ovarian cancer. Cancer Cell Int. 2022 Dec 30;22(1):422. doi: 10.1186/s12935-022-02838-x. PMID: 36585738; PMCID: PMC9805260.
- Murphy SK, Berchuck A, Whitaker R, Sfakianos G, Huang Z. Primary and recurrent (second-look surgery) serous epithelial ovarian cancers Illumina Infinium HumanMethylation450 BeadChip data. Duke Research Data Repository. 2021.
- Murphy S, Berchuck A, Whitaker R, Sfakianos G, Huang Z. Gene Expression using Affymetrix Human Genome U133 Plus 2 Arrays from 16 Primary and Recurrent Serous Epithelial Ovarian Cancers. Duke Research Data Repository. 2021.
- Berchuck A, Iversen ES, Lancaster JM, Pittman J, Luo J, Lee P, Murphy S, Dressman HK, Febbo PG, West M, Nevins JR, Marks JR. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 2005 May 15;11(10):3686-96. doi: 10.1158/1078-0432.CCR-04-2398. PMID: 15897565.
- Langdon SP, Lawrie SS. Establishment of ovarian cancer cell lines. Methods Mol Med. 2001;39:155-9. doi: 10.1385/1-59259-071-3:155. PMID: 21340766.
- Johnson SW, Laub PB, Beesley JS, Ozols RF, Hamilton TC. Increased platinum-DNA damage tolerance is associated with cisplatin resistance and cross-resistance to various chemotherapeutic agents in unrelated human ovarian cancer cell lines. Cancer Res. 1997 Mar 1;57(5):850-6. PMID: 9041185.
- Lee HR, Leslie F, Azarin SM. A facile in vitro platform to study cancer cell dormancy under hypoxic microenvironments using CoCl2. J Biol Eng. 2018 Aug 3;12:12. doi: 10.1186/s13036-018-0106-7. PMID: 30127847; PMCID: PMC6091074.
- Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016 Jul 8;44(W1):W147-53. doi: 10.1093/nar/gkw419. Epub 2016 May 17. PMID: 27190236; PMCID: PMC4987948.
- Kim J, Park EY, Kim O, Schilder JM, Coffey DM, Cho CH, Bast RC Jr. Cell Origins of High-Grade Serous Ovarian Cancer. Cancers (Basel). 2018 Nov 12;10(11):433. doi: 10.3390/cancers10110433. PMID: 30424539; PMCID: PMC6267333.
- Aslani FS, Maleknasab M, Akbarzadeh-Jahromi M. Fallopian Tube Epithelial Changes in Ovarian Serous Tumors Compared with Control Group: A Single-Center Study. Niger Med J. 2019 Mar-Apr;60(2):47-52. doi: 10.4103/nmj.NMJ_27_19. PMID: 31462842; PMCID: PMC6688397.
- Kagohara LT, Stein-O'Brien GL, Kelley D, Flam E, Wick HC, Danilova LV, Easwaran H, Favorov AV, Qian J, Gaykalova DA, Fertig EJ. Epigenetic regulation of gene expression in cancer: techniques, resources and analysis. Brief Funct Genomics. 2018 Jan 1;17(1):49-63. doi: 10.1093/bfgp/elx018. PMID: 28968850; PMCID: PMC5860551.
- Zhang Y, Mei Q, Liu Y, Li X, Brock MV, Chen M, Dong L, Shi L, Wang Y, Guo M, Nie J, Han W. The safety, efficacy, and treatment outcomes of a combination of low-dose decitabine treatment in patients with recurrent ovarian cancer. Oncoimmunology. 2017 May 17;6(9):e1323619. doi: 10.1080/2162402X.2017.1323619. PMID: 28932630; PMCID: PMC5599090.
- Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting Tumor Microenvironment for Cancer Therapy. Int J Mol Sci. 2019 Feb 15;20(4):840. doi: 10.3390/ijms20040840. PMID: 30781344; PMCID: PMC6413095.
- Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008 Oct 6;27(45):5904-12. doi: 10.1038/onc.2008.271. PMID: 18836471; PMCID: PMC3689267.
- Koontongkaew S, Amornphimoltham P, Monthanpisut P, Saensuk T, Leelakriangsak M. Fibroblasts and extracellular matrix differently modulate MMP activation by primary and metastatic head and neck cancer cells. Med Oncol. 2012 Jun;29(2):690-703. doi: 10.1007/s12032-011-9871-6. Epub 2011 Mar 6. PMID: 21380786.
- Das A, Monteiro M, Barai A, Kumar S, Sen S. MMP proteolytic activity regulates cancer invasiveness by modulating integrins. Sci Rep. 2017 Oct 27;7(1):14219. doi: 10.1038/s41598-017-14340-w. PMID: 29079818; PMCID: PMC5660204.
- Hornebeck W, Emonard H, Monboisse JC, Bellon G. Matrix-directed regulation of pericellular proteolysis and tumor progression. Semin Cancer Biol. 2002 Jun;12(3):231-41. doi: 10.1016/s1044-579x(02)00026-3. PMID: 12083853.
- Nii T, Kuwahara T, Makino K, Tabata Y. A Co-Culture System of Three-Dimensional Tumor-Associated Macrophages and Three-Dimensional Cancer-Associated Fibroblasts Combined with Biomolecule Release for Cancer Cell Migration. Tissue Eng Part A. 2020 Dec;26(23-24):1272-1282. doi: 10.1089/ten.TEA.2020.0095. Epub 2020 Jun 26. PMID: 32434426.
- Senthebane DA, Rowe A, Thomford NE, Shipanga H, Munro D, Mazeedi MAMA, Almazyadi HAM, Kallmeyer K, Dandara C, Pepper MS, Parker MI, Dzobo K. The Role of Tumor Microenvironment in Chemoresistance: To Survive, Keep Your Enemies Closer. Int J Mol Sci. 2017 Jul 21;18(7):1586. doi: 10.3390/ijms18071586. PMID: 28754000; PMCID: PMC5536073.
- Wu Y, Sarkissyan M, Vadgama JV. Epigenetics in breast and prostate cancer. Methods Mol Biol. 2015;1238:425-66. doi: 10.1007/978-1-4939-1804-1_23. PMID: 25421674; PMCID: PMC4364390.
- Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, Han J, Wei X. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019 Dec 17;4:62. doi: 10.1038/s41392-019-0095-0. PMID: 31871779; PMCID: PMC6915746.
- Alkasalias T, Moyano-Galceran L, Arsenian-Henriksson M, Lehti K. Fibroblasts in the Tumor Microenvironment: Shield or Spear? Int J Mol Sci. 2018 May 21;19(5):1532. doi: 10.3390/ijms19051532. PMID: 29883428; PMCID: PMC5983719.
- Westermarck J, Kähäri VM. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999 May;13(8):781-92. PMID: 10224222.
- Kähäri Nr. aV. Matrix Metalloproteinases in Cancer Cell Invasion. Landes Bioscience. 2000-2013.
- He X, Dai J, Fan Y, Zhang C, Zhao X. Regulation function of MMP-1 downregulated by siRNA on migration of heat-denatured dermal fibroblasts. Bioengineered. 2017 Nov 2;8(6):686-692. doi: 10.1080/21655979.2016.1267885. Epub 2017 Feb 22. Retraction in: Bioengineered. 2020 Dec;11(1):558. PMID: 28277161; PMCID: PMC5736340.
- Jabłońska-Trypuć A, Matejczyk M, Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. 2016;31(sup1):177-183. doi: 10.3109/14756366.2016.1161620. Epub 2016 Mar 30. PMID: 27028474.
- Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002 Mar;2(3):161-74. doi: 10.1038/nrc745. PMID: 11990853.
- Juncker-Jensen A, Deryugina EI, Rimann I, Zajac E, Kupriyanova TA, Engelholm LH, Quigley JP. Tumor MMP-1 activates endothelial PAR1 to facilitate vascular intravasation and metastatic dissemination. Cancer Res. 2013 Jul 15;73(14):4196-211. doi: 10.1158/0008-5472.CAN-12-4495. Epub 2013 May 16. PMID: 23687338; PMCID: PMC3754905.
- Quintero-Fabián S, Arreola R, Becerril-Villanueva E, Torres-Romero JC, Arana-Argáez V, Lara-Riegos J, Ramírez-Camacho MA, Alvarez-Sánchez ME. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front Oncol. 2019 Dec 6;9:1370. doi: 10.3389/fonc.2019.01370. PMID: 31921634; PMCID: PMC6915110.
- A knowledge resource on cell lines. https://www.cellosaurus.org. Dec 2022.
- Laios A, Mohamed BM, Kelly L, Flavin R, Finn S, McEvoy L, Gallagher M, Martin C, Sheils O, Ring M, Davies A, Lawson M, Gleeson N, D'Arcy T, d'Adhemar C, Norris L, Langhe R, Saadeh FA, O'Leary JJ, O'Toole SA. Pre-Treatment of platinum resistant ovarian cancer cells with an MMP-9/MMP-2 inhibitor prior to cisplatin enhances cytotoxicity as determined by high content screening. Int J Mol Sci. 2013 Jan 22;14(1):2085-103. doi: 10.3390/ijms14012085. PMID: 23340649; PMCID: PMC3565367.
- Ojalvo LS, Thompson ED, Wang TL, Meeker AK, Shih IM, Fader AN, Cimino-Mathews A, Emens LA. Tumor-associated macrophages and the tumor immune microenvironment of primary and recurrent epithelial ovarian cancer. Hum Pathol. 2018 Apr;74:135-147. doi: 10.1016/j.humpath.2017.12.010. Epub 2017 Dec 27. PMID: 29288043.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
