Global Journal of Obesity, Diabetes and Metabolic Syndrome
Novel key gene tenascin C related to extracellular matrix accumulation in diabetic nephropathy kidney tubules: Results of integrative bioinformatics analysis
Liang Zhang1, Zheng Wang1, Manrong He1, Yongdi Zuo1, Jun Li2,3 and Wanxin Tang1*
2The Center of Gerontology and Geriatrics, West China Hospital, Sichuan University, Chengdu, Sichuan, China
3National Clinical Research Center of Geriatrics, West China Hospital, Sichuan University, Chengdu, Sichuan, China
Cite this as
Zhang L, Wang Z, He M, Zuo Y, Tang W, et al. (2020) Novel key gene tenascin C related to extracellular matrix accumulation in diabetic nephropathy kidney tubules: Results of integrative bioinformatics analysis. Glob J Obes Diabetes Metab Syndr 7(2): 042-052. DOI: 10.17352/2455-8583.000047Background: As the incidence of type 2 diabetes increases year by year, the number of individuals diagnosed with Diabetic Nephropathy (DN) has increased steeply. DN is characterized by glomerular sclerosis, tubulointerstitial fibrosis and atrophy. However, most of the previous studies on the pathogenesis of DN were focused on glomeruli, and now more and more evidences show that tubulointerstitial fibrosis plays an important role in the progress of DN. Bioinformatics analysis can be used in discovering disease-causing genes, biomarkers, and therapeutic targets through global analysis. In this study, we used this method to find and verify novel genes in DN.
Materials and methods: Microarray expression levels were downloaded from Gene Expression Omnibus datasets. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were conducted after Differentially Expressed Genes (DEGs) were identified. Hub genes were filtered on the basis of the result of GO enrichment, and functional analysis was performed by browsing the GeneCards website and the latest literatures. The expression levels of all the hub genes in HK2 cells stimulated by high glucose were verified, and TeNascin C (TNC), which is important and interesting, was verified through animal experiment.
Result: The relative expression levels of 54 samples were obtained, and 382 DEGs were identified. 286 GO terms and 100 KEGG pathways were enriched remarkably. Nine of the key genes of interest: MMP7, TNC, LUM, LTF, IGLC1, LYZ, CXCL6, CYP27B1 and FOS were selected and verified in HK2 cells stimulated by high glucose. quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) indicated that the mRNA levels of most of the hub genes were up-regulated and had an accordance rate of approximately 66.7%. In addition, the expression of TNC in renal tubular tissue of STZ-induced diabetic nephropathy rat gradually increased with time.
Conclusion: Several novel key genes were discovered and verified in this study, and TNC plays an important role in the renal fibrosis of DN and is expected to be a new therapeutic target. This study provides a theoretical basis and data resources for further research.
Abbreviations
DN: Diabetic Nephropathy; ESRD: End-Stage Renal Disease; ECM: ExtraCellular Matrix; TGF-β: Transforming Growth Factor Beta; MMP9: Matrix MetalloPeptidase 9; APC: Adenomatous Polyposis Coli; GEO: Gene Expression Omnibus; DEGs: Differentially Expressed Genes; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; NCBI: National Center for Biotechnology Information; Limma: Liner models for microarray data; FC: Fold Change; CC: Cellular Component; MF: Molecular Function; BP: Biological Process; HK2: Human Kidney 2; qRT-PCR: quantitative Real-Time Polymerase Chain Reaction; CT: Cycle Threshold; PBS: Phosphate-Buffered Saline; MMP7: Matrix MetalloPeptidase 7; TNC: TeNascin C; LUM: LUMican; LTF: LacTotransFerrin; IGLC1: ImmunoGlobulin Lambda Constant 1; LYZ: LYsoZyme; (CXCL6): Chemokine (C-X-C motif) Ligand 6; CYP27B1; CYtochrome P450 family 27 subfamily B member 1; ALB: ALBumin; C3: Complement component 3; IOD: Integrated Optical Density
Background
The incidence of diabetes, especially type 2 diabetes continues to increase globally. In 2015, approximately 415 million people have diabetes, and It is estimated that the number of diabetes will rise to 642 million by 2040 [1]. As we all know, diabetes (both type 1 and type 2) can cause various complications such as diabetic retinopathy, diabetic peripheral vascular disease, and diabetic autonomic neuropathy. What`s more, Diabetic Nephropathy (DN) is also one of the main complications of diabetes, and it is a major microvascular complication caused by uncontrolled high blood glucose for a long time. DN is characterized by proteinuria and progressive reduction in kidney function. The number of individuals diagnosed with DN has increased sharply owing to the high incidence rate of type 2 diabetes [2-4]. About 10% of deaths in type 2 diabetes patients were attributed to renal failure, and in the United States End-Stage Renal Disease (ESRD) caused by diabetes accounts for as high as 44% [4], even else, in some western countries, DN has become the most common cause of ESRD [5,6].
However, the pathogenesis of DN is extremely complex and has not been fully understood as yet. DN is characterized by glomerulosclerosis, tubulointerstitial fibrosis, and atrophy [7] and most of previous studies on the mechanism of DN are focused on glomeruli. Growing evidence reveals that the tubulointerstitial fibrosis plays an important role in the progression of DN [8], and the accumulation of ExtraCellular Matrix (ECM) is one of the most important mechanisms of tubulointerstitial fibrosis [9].
Current studies suggest that Transforming Growth Factor beta (TGF-β) is involved in the progression of tubulointerstitial fibrosis in DN [10,11]. Matrix MetalloPeptidase 9 (MMP9) [12-14], and Adenomatous Polyposis Coli (APC) [15] et al. may also play a role in the progression of tubulointerstitial fibrosis in DN. However, these scattered studies cannot provide a global impression for DN, because of the complexity of DN`s mechanism. In addition, numerous genes and targets have not been discovered.
Bioinformatics analysis surfaces with the emergence of massive gene data, and is a powerful methodology that can be used in discovering disease-causing genes, biomarkers, and therapeutic targets through global analysis. For example, using bioinformatics analysis methods, Gurudeeban Selvaraj, et al. [16] identified that mitogen-activated protein kinase 1 and aurora kinase are the hub nodes and identified candidate drugs in lung adenocarcinoma. However, the application of bioinformatics analysis methods in kidney diseases has just started. Cui Y, et al. [17] identified new biomarkers and therapeutic targets for immunoglobulin A nephropathy and hypertensive nephropathy. To date, corresponding studies in the field of DN, particularly of the kidney tubule, remain scarce.
In this study, we integrated the data of several different platforms about kidney tubule in DN from the Gene Expression Omnibus (GEO) dataset through bioinformatics methods and identified Differentially Expressed Genes (DEGs). We then conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses and filtered hub genes through the network between the GO terms and DEGs. Finally, we performed in vivo and in vitro experiments to verify the novel hub genes.
Materials and Methods
Microarray data
The microarray expression data of GSE30529, GSE99325, GSE35487, and GSE37455 [18-21] were downloaded from GEO datasets (http://www.ncbi.nlm.nih.gov/geo/) in National Center for Biotechnology Information (NCBI). GSE30529 was obtained from the GPL571 platform: [HG-U133A_2], GSE99325 was obtained from the GPL19109 platform: [HG-U133_Plus_2] and GPL19184 platform: [HG-U133A],GSE35487 was obtained from the GPL96 platform: [HG-U133A], and GSE37455 was obtained from the GPL11670 platform: [Hs133P_Hs_ENTREZG.cdf] and GPL14663 platform: [Affy_HGU133A_CDF_ENTREZG_10]. Fifty-four samples, consisting of 28 diabetic human kidney tubule samples and 26 control human kidney tubule samples, were acquired for further gene profile analysis.
Data preprocessing
The raw data of GSE30529, GSE99325, GSE35487, and GSE37455 in CEL files were converted into a probe expression matrix by the R Affy package [22]. Background correction, normalization, and log-transformation were subsequently conducted by the ComBat function and Bayesian analysis of the surrogate variable analysis package in R [23] for the removal of the interval differences.
Identification of DEGs
Liner models for microarray data (Limma) in R [24] was used in identifying DEGs. Adjusted P values of <0.05 and log FC of ≥1 were set as the cutoff criteria, and the results were illustrated with a volcano plot. Bidirectional hierarchical clustering analysis was performed on the first 30 up-regulated and 30 down-regulated DEGs with the pheatmap package (version 1.0.8) in R [25] for the visualization of the differences between the DEGs of the DN and normal samples.
GO enrichment analysis
GO analysis [26] is a powerful bioinformatics tool for annotating the biological functions behind a large list of genes. GO mainly contains three categories: Cellular Component (CC), Molecular Function (MF), and Biological Process (BP). GO analysis was performed on the basis of the DEGs by using the gseGO function in clusterProfiler (version 3.5) [27], and the adjusted P value of < 0.05 was set as the cutoff criterion. The top 10 GO terms of each category ranked by an adjusted P value were visualized through bubble plot by the GO plot package in R [28]. The connections between the most significant GO terms in BP and participating genes were also visualized using the UpSet plot package in R [29].
KEGG pathway enrichment analysis
KEGG [30] is a pathway-related database that provides the information about genomes, biological pathways, diseases, and chemical substances. The gseKEGG function in clusterProfiler was used in the enrichment analysis of KEGG, which was performed on the basis of all the genes obtained from the primary data, and the analysis results were visualized with dot plot by the enrichplot package in R.
Hub gene analysis
The network between DEGs and the top five GO terms in each category drawn by GOplot package in R can reveal the connection between genes and GO terms and the fold change of each gene expressed by color depth. The DEGs that had considerably large fold change and considerably small P value and were enriched in the top GO terms were considered hub genes. Therefore, the genes that were considered hub genes were evident from the network. Only the top DEGs from the top five GO terms in BP were selected and further subjected to functional analysis because of the importance of BP in disease and our limited energy. The other DEGs were also valuable. We browsed the detailed introductions and functions of each hub gene in the GeneCards website (https://www.genecards.org), which is a comprehensive and authoritative compendium of annotative information about human genes [31]. We then reviewed the latest literature to determine whether these genes are involved in the mechanism of DN. Furthermore, the DEGs were validated by subsequent in vitro and in vivo experiments.
Cell culture
Human renal tubular epithelial cells (Human Kidney 2, HK2) were bought from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle medium/F12 (Hyclone, NY, USA) with additional 10% fetal bovine serum (Hyclone, NY, USA) in a humidified incubator at 37 °C under 5% CO2. All of the cells were cultured in a complete medium with the same glucose concentration at 80% confluence after synchronization. The cells were lysed at 0, 12, 24, and 48 h for subsequent experiments.
qRT-PCR(quantitative Real-Time Polymerase Chain Reaction)
Cells or grand tissues were lysed with TRIzol-LS (Ambion), and total mRNA was prepared with RNeasy Plus Mini Kit (BioTeke RP1202) according to the manufacturer’s instructions. Genome removal reaction was performed using DNA Eraser and PrimeScript RT Reagent Kit(RR047A) was used for the acquisition of cDNA through reverse transcription. A qRT-PCR system (PIKORed 96, ThermoFisher) was subsequently used for PCR analysis for 45 cycles under the following conditions: 95 °C for 35 s,55 °C for 30 s, and 72 °C for 30 s. β-Actin was selected as internal control, and the primer sequences used for PCR are listed in Table 1. The Thermo Scientific PikoReal software (Thermo) was used in analyzing the Cycle Threshold (CT) value of each test sample in the PCR process. We calculated the relative mRNA expression level through 2-△CT, representing the expression fold of genes in comparison with β-actin.
Animal model construction
Male Sprague-Dawley rats (8 weeks old, 200–250 g) were purchased from the Dashuo Experimental Animal Center (Chengdu, China). All animals were fed under the same conditions for 7 days before the experiments. A total of 24 rats were injected with a single dose of streptozotocin (65 mg/kg, intraperitoneally, Sigma-Aldrich, Saint Louis, Missouri, USA), and six rats, as control (0-week), were injected with the same dose of normal saline. The diabetes model was confirmed by detecting blood glucose levels of >16.7 mmol/L from the random samples for 3 days. Then, 24 diabetes model rats were randomly and evenly divided into four groups: 4-weeks, 8-weeks, 12-weeks, and 16-weeks. Finally, all the rats were euthanized at the corresponding time, and kidney tissues were obtained. The animal studies were followed the guidelines issued by the National Institute of Health Guide for the Care and Use of Laboratory Animals. The protocol was approved by the Animal Care Committee of West China Hospital, Sichuan University, China.
Immunohistochemistry
Kidney tissue sections were deparaffinized, hydrated, and then washed with Phosphate-Buffered Saline (PBS). Endogenous peroxidase activity was quenched by incubating the sections in methanol solution containing 3% H2O2. The sections were then incubated overnight with primary antibodies at 4 °C and then washed with PBS. Secondary antibody was added and then reacted at room temperature for 30 min. The slides were washed with PBS, then incubated with diaminobenzidine tetrahydrochloride, and finally counterstained with hematoxylin. The positive region was observed under a positive fluorescent microscope (Olympus, Japan) and photographed under 400× magnification. Image-Pro Plus software (version 6.0) was used in calculating the positive region proportion.
Statistical analysis
Linear model fitting and Bayesian test in R (version 3.5.1) were used in identifying DEGs, Fisher`s exact test in R (version 3.5.1) was used in GO and KEGG enrichment analysis, and false discovery rate of <0.05 was considered to be statistically significant. With SPSS 22.0, t-test was performed on the final data of qRT-PCR and immunohistochemistry semi-quantitative analysis, and p value of <0.05 was considered to be statistically significant.
Results
Data preprocessing and identification of DEGs
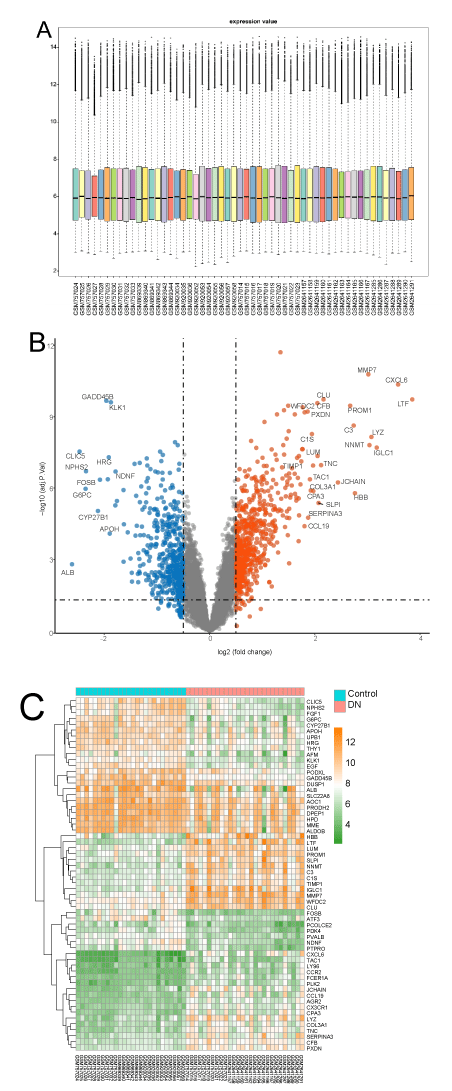
After background correction, normalization, and log-transformation, the relative expression levels of the 54 samples were obtained (Figure 1a). The straight middle line and the uniform distribution indicated the high quality and comparability of the data in our study. On the basis of the criteria that we initially set, 382 DEGs were identified in diabetic human kidney tubule samples compared with the control. The volcanic plot of DEGs (Figure 1b) shows the criteria and results of DEGs. Most of the DEGs were up-regulated. The heatmap (Figure 1c), result of hierarchical clustering analysis displayed the gene expression levels of the top 30 DEGs in each sample. Table 2 shows the 15 up-regulated and 15 down-regulated DEGs with the largest log (FC) values. Log (FC), average expression level, adjusted P value, and expression pattern are also indicated in the table.
Enrichment analysis
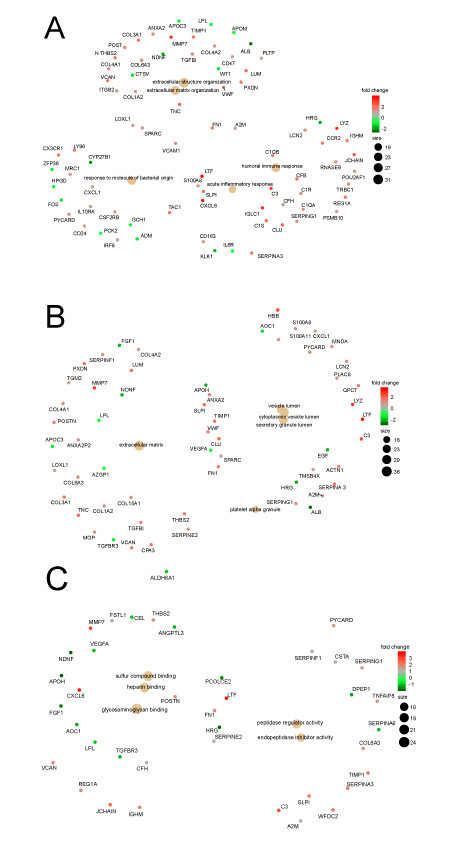
A total of 286 GO terms were found to be significantly enriched in the DEGs, including 201 in BP, 44 in CC, and 41 in MF, and the top 10 GO terms of each category ranked by adjusted P value, are shown in Figure 2a. The list of GO terms illustrated that the “extracellular structure organization,” and “ECM organization,” in BP, and the “ECM,” “proteinaceous ECM,” and “ECM component,” in CC were significantly enriched. Basing on this finding, we speculate that change in ECM might lead to tubulointerstitial fibrosis and plays an important role in the mechanism of DN. Thus, the DEGs contained by all of these GO terms may play a key role in the pathogenesis of DN. Several other GO terms related to immune response and endopeptidase also possibly contribute to the pathogenesis of DN. The connection between the top 10 GO terms in BP and the participating genes is shown in Figure 2b. Most of the DEGs were associated with multiple GO terms. Even else, one DEG simultaneously participated in seven GO terms, so this special DEG was the hub gene and the potential therapy target.
A total of 100 KEGG pathways, consisting 65 activated and 35 suppressed pathways, were enriched considerably and the top 12 of each regulation type are shown in Figure 2c and Table 3. Most of the enriched KEGGs were related to energy metabolism and contained carbohydrates, fats, and amino acids, such as “citrate cycle,” “butanoate metabolism,” and “tryptophan metabolism,” and all of these KEGGs were suppressed. The inhibited energy metabolism probably leads to the occurrence and development of DN, but the specific mechanism is unknown. Certain pathways related to immune response, such as “phaGO termsome,” “natural killer cell mediated cytotoxicity,” and “chemokine signaling pathway,” were also enriched remarkably. This result showed the important role of immunity in DN.
Hub-genes analysis
As shown in Figure 3, GO terms were closely linked with one another through multiple hub genes. Only the top two genes in GO terms (up-regulated and down regulated) and |log(FC)|>2, were set as the screening criteria. Thus, DEGs, including Matrix MetalloPeptidase 7 (MMP7), TeNascin C (TNC), LUMican (LUM), LacTotransFerrin (LTF), ImmunoGlobulin Lambda Constant 1 (IGLC1), lysozyme (LYZ), Chemokine (C-X-C motif) ligand 6 (CXCL6), CYtochrome P450 family 27 subfamily B member 1 (CYP27B1), and FOS, were selected for further analysis. Only CYP27B1 and FOS were down-regulated. The functions and roles of ALBumin (ALB) and Complement component 3 (C3) had been clarified; thus, ALB and C3 were not included in the subsequent analysis despite their significant enrichment and large fold change. By reviewing the latest literature, we found that MMP7, LUM, CXCL6, CYP27B1, and FOS were associated with DN. However, TNC, LTF, IGLC1, and LYZ were not reported. This result proved that our analysis results were credible and could provide new research directions. The detailed functions of these hub-genes are listed in Table 4. All of the hub-genes were subjected to subsequent in vitro validation experiments. TNC, one of the hub genes, is novel for DN and encodes an ECM protein with a spatially and temporally restricted tissue distribution. The result of GO analysis showed that ECM was closely related to DN. Thus, TNC attracted our attention and interest and was subjected to animal verification experiment.
qRT-PCR
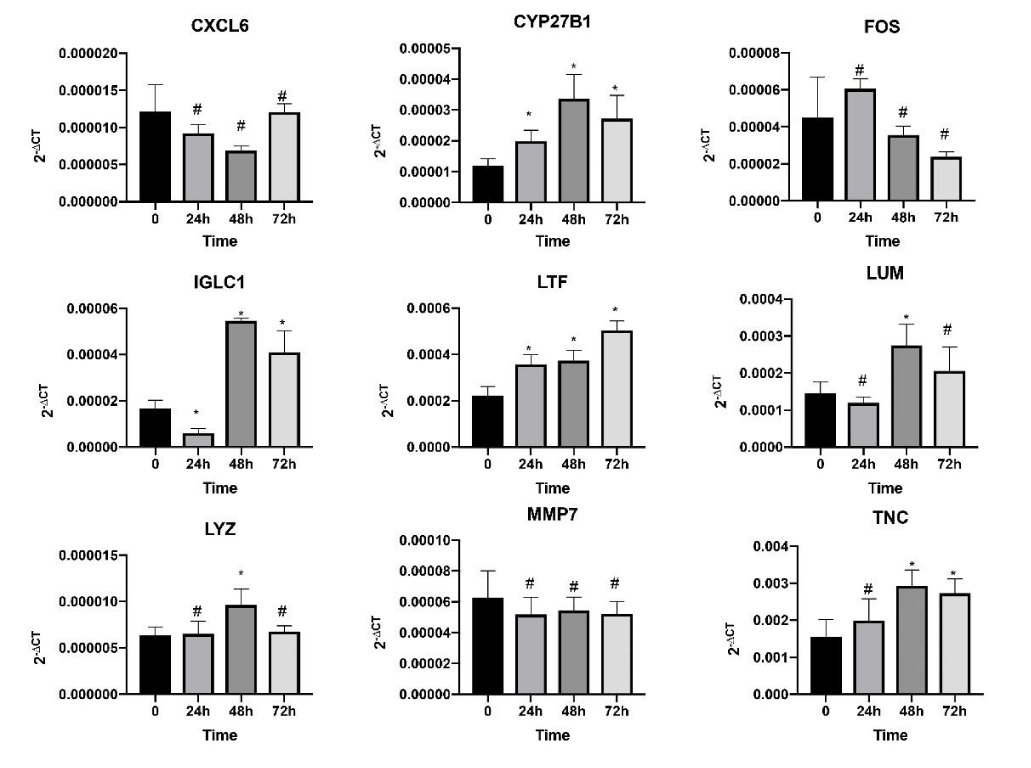
We validated the nine hub genes expression levels in the HK2 cells in vitro. qRT-PCR indicated that the mRNA levels of TNC, LUM, LTF, IGLC1, LYZ, and CYP27B1 were up-regulated after being stimulated by high glucose. Simultaneously, FOS was down-regulated, but its change had no statistical difference. In addition, the change in MMP7 and CXCL6 were not substantial. Changes in all of the other gene expression levels except those of CYP27B1, MMP7, and CXCL6 were consistent with the bioinformatics analysis results, and the accordance rate was approximately 66.7%. The expression levels represented by 2-∆CT of all the genes and the time are shown in Figure 4. Moreover, the result demonstrated that the expression levels of these genes stimulated by high glucose were time dependent. The expression levels of CYP27B1 and TNC were increased in all time points and had the maximum expression values at 48 h. The expression levels of IGLC1 and LUM were increased only at 48 and 72 h and were inhibited at 24 h. Furthermore, LYZ had increased expression at 48 h only.
Immunohistochemistry
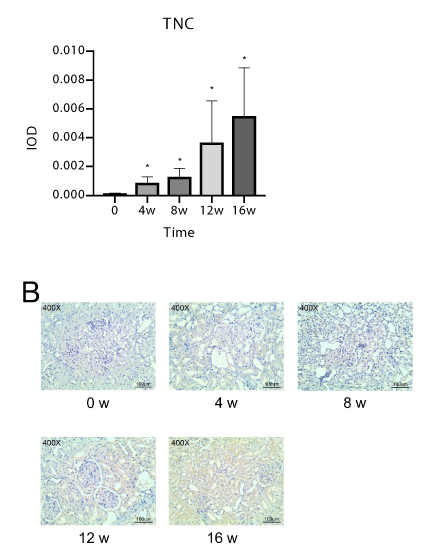
We obtained 20 photos of satisfactory quality. Figure 5a shows the average integrated optical density at each time point, which represents the expression of the TNC gene. The rats were established as a DN model. The expression of TNC in their kidney tissues gradually increased with time, and the difference was statistically significant compared with the 0-week group. Certain representative images at each time point are shown in Figure 5b. The result of the animal validation experiment showed that the expression of TNC, which is involved in the final formation of renal fibrosis gradually increased with the duration of DN increases. In addition, the expression levels of TNC were consistent with the results of the bioinformatics analysis and in vitro validation experiment.
Discussion
It is currently known that glomerular endothelial cells, glomerular mesangial cells, podocytes and renal tubular epithelial cells of the kidney are the targets of hyperglycemia injury. Glomerular sclerosis, thickening of the glomerular basement membrane, glomerular hypertrophy, mesangial cell expansion, podocyte loss, renal cell hypertrophy and tubular interstitial fibrosis are the main pathological changes that occur during DN [37], and which eventually leads to progressive albuminuria. Because the pathogenesis of DN is extremely complicated, it has not yet been elucidated. Early studies found that advanced glycosylation end products, polyol pathway activation, protein kinase activity, abnormal lipid metabolism and hemodynamic changes are related to the occurrence and development of DN. However, these scattered studies cannot provide an overall impression of the pathogenesis of DN, and the biomarkers for early diagnosis and effective treatment are still lacking nowadays. At the same time, there are also a large number of pathogenic genes and potential treatment targets have not been found. Therefore, the pathogenesis of diabetic nephropathy needs to be further studied.
GEO is a public dataset that contains a large amount of array- and sequence-based data. Each raw data can provide scientific and large-scale information for the comprehensive implementation of bioinformatics analysis. Bioinformatics analysis is a powerful methodology, that is objectively, advanced, and globally. And the results of it are reliable. Our study is the first to research the tubule cells of DN through bioinformatics analysis. By obtaining each sample from DN individually, changes in disease-causing genes were directly determined. In addition, we increased the sample size, acquired and integrated all relevant data, and found a large number of important DEGs. This study provides useful resources for in-depth research. Furthermore, the results of our bioinformatics analysis have been validated in vivo and in vitro using appropriate and scientific methods, making our findings credible and reproducible.
In order to further analyze the function of DEGs and explore the molecular mechanism of DN, we performed the functional enrichment analysis of GO and KEGG based on DEGs. A total of 286 GO terms were found to be significantly enriched, and “Extracellular structure organization” is the most significant GO term filtered by enrichment analysis. Several GO terms related to ECM, such as “ECM organization,” “ECM,” “proteinaceous EMC,” and “ECM component,” were also enriched significantly. This result indicated that ECM plays an important role in the occurrence and development of DN. A published study also showed that the excessive synthesis and decreased breakdown of the ECM lead to glomerulosclerosis and tubulointerstitial fibrosis, which further lead to the development of ESRD [38]. However, the mechanism between ECM and glomerulosclerosis is extremely intricate. Meng X.M, et al. [39] proposed that TGF-β is the master regulator of fibrosis. Moreover, the MMP family and TIMP1 are also associated with fibrosis [13]. This finding further confirms that our study is consistent with previous research.
What`s more, several other GO terms related to immune response and endopeptidase were also enriched significantly, such as “humoral immune response”, “acute inflammatory response”, “response to interferon-gamma” and “negative regulation of endopeptidase activity”. Early studies have already made it clear that immune inflammation is the most important factor that promotes podocyte damage and albuminuria in DN, and IL-6, IL-8 and TNF-a are all significantly increased in DN [40]. After IL-6 binds to IL-6R, it binds to the gp130 signal transduction chain, which can lead to autophosphorylation and changes in conformational structure of Janus kinase, thereby inducing intracellular signals and activating transcription factors, signal transducers and transcription activators -3, and eventually lead to the growth and proliferation of mesangial cells [41]. In addition, more and more evidences also indicate that the complement system has a pathogenic role in the development of diabetic nephropathy, and it is believed that inhibiting specific components of the complement system may be an effective treatment strategy for the treatment of diabetic nephropathy [42]. These studies have further confirmed that our research is consistent with previous studies.
As the result of enrichment analysis of KEGG pathways, “citrate cycle”, “butanoate metabolism” and “tryptophan metabolism” and several other significantly enriched KEGG are related to the metabolism of glucose, fat and amino acid, and these pathways are inhibited. Mitochondria are the place of energy metabolism and mitochondrial dysfunction can lead to an increase in reactive oxygen species and aggravate the damage of oxidative stress to vascular endothelial cells and podocytes [43]. Manjula Darshi, et al. through the study of DN metabolomics also found that mitochondrial function and fatty acid oxidation play a key role in the occurrence and development of DN [44]. Therefore, we infer that the inhibition of energy metabolism may be involved in the occurrence and development of DN, but the more specific mechanisms need to be further studied, and it also provides us with new research directions.
Through our previous data analysis, a number of key genes were screened out and verified in HK2 cells stimulated by high glucose. Among them, TNC, LTF, IGLC1 and LYZ are currently related and there is no literature report whether they are related to DN. LTF, encoding lactoferrin, has the regulation of iron absorption, the regulation of immune response, as well as antimicrobial, antiviral, antifungal and antiparasitic activities [45]. In this study, the results of bioinformatics analysis and in vitro experiment have shown that the high expression of LTF in the renal tubule tissue of patients with diabetic nephropathy may be involved in the damage to the kidney along with other inflammation and immune-related pathways. IGLC1, which encodes the constant region of the immunoglobulin light chain, and participates in the effect phase of humoral immunity. In our verification experiment, we see that IGLC1 is significantly highly expressed at 48 hours and 72 hours, and on the contrary is low at 24 hours. As we all know, blood system diseases such as multiple myeloma can cause renal function decline due to the deposition of monoclonal globulin or light chain in the kidney. In DN, is it possible that the high expression of IGLC1 increases the load of light chain on the kidney and aggravate kidney damage? This aspect has once again provided us with new research directions. LYZ, which encodes human lysozyme, has antibacterial activity in many kinds of bacteria, but its function lacks specificity, and there are few related researches. Although there is no clear evidence to proves that these genes are related to DN, they are all related to immunity, which plays a key role in the pathogenesis of DN, and is expected to become a new research direction.
TNC is a homohexamer that contains multiple epidermal growth factor-like and fibronectin type-III domains linked by disulfides and can present several molecular forms through alternative splicing and protein modifications. This multimodal structure allows tenascins to interact with a large number of highly diverse ligands (provided by RefSeq, July 2011). Another notable feature of TNC is its tightly controlled pattern of gene expression. Few TNCs are found in adults except in stem cell niches, tendons, inflammation and trauma sites, and the stroma of solid tumors [46,47]. In addition, TNC has an exceedingly wide range of functions, and the adhesion-regulating ECM protein is probably well-known because of its ability to link to fibronectin and change cell proliferation and signaling mediated through fibronectin and integrins [48]. Finally, TNC is associated with certain diseases and has the opportunity to be used in diagnosis and treatment. For example, the detection of high levels of TNC in body fluids may have a diagnostic value for diseases, including heart disease, rheumatoid arthritis, and cancer [49,50]. Moreover, TNC may be a good target for adjuvant chemotherapy in breast cancer [51]. In our study, the gene expression analysis of human renal tubule cells in the in vitro and vivo experiments indicated that TNC was highly expressed in the DN group. Previous research showed that a TNC-enriched microenvironment is associated with kidney fibro genesis [52]. Therefore, TNC is involved in the pathogenesis of DN, and its possible pathogenic mechanism increases the synthesis of the ECM by adhesion-regulating ECM protein and leads to glomerulosclerosis and tubulointerstitial fibrosis.
However, the samples that we have analyzed are from patients with DN at different stages because of the limited samples, which may not reflect the progression of the disease. This limitation may be the deficiency of this study, but our validation experiment supplements and enhances the credibility of the analysis results.
Conclusions
This study is the first to use bioinformatics analysis for identifying several novel key genes in tubule cells in DN. TNC, a novel gene of DN, plays an important role in the renal fibrosis of DN, and is expected to be a new therapeutic target. The results provide a theoretical basis and data resources for further research. This study also provides clues for finding new biomarkers and therapeuti targets.
Supported by the Foundation of Science and Technology Department of Sichuan Province (grant number 2018SZ0378 and 2018SZ0175).
- International Diabetes Federation (2015) IDF Diabetes Atlas, Seventh Edition. Diabetes Atlas. Link: https://bit.ly/32cTmMQ
- Koye DN, Magliano DJ, Nelson RG, Pavkov ME (2018) The Global Epidemiology of Diabetes and Kidney Disease. Adv Chronic Kidney Dis 25: 121-132. Link: https://bit.ly/2QgJ9cQ
- Ingelfinger JR, Jarcho JA (2017) Increase in the Incidence of Diabetes and Its Implications. N Engl J Med 376: 1473-1474. Link: https://bit.ly/32gZeoE
- Zheng Y, Ley SH, Hu FB (2018) Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol 14: 88-98. Link: https://bit.ly/32j20JY
- US Renal Data System 2019 Annual Data Report: Epidemiology of Kidney Disease in the United States. Am J Kidney Dis . Link: https://bit.ly/34teBNk
- Sharma S, Sarnak MJ (2017) Epidemiology: The global burden of reduced GFR: ESRD, CVD and mortality. Nat Rev Nephrol 13: 447-448. Link: https://bit.ly/2Yti6iU
- Qi C, Mao X, Zhang Z, Wu H (2017) Classification and Differential Diagnosis of Diabetic Nephropathy. J Diabetes Res 2017: 8637138. Link:
- Fan Y, Yi Z, D'Agati VD, Sun Z, Zhong F, et al. (2019) Comparison of Kidney Transcriptomic Profiles of Early and Advanced Diabetic Nephropathy Reveals Potential New Mechanisms for Disease Progression. Diabetes 68: 2301-2314. Link: https://bit.ly/2Yt2s7i
- Humphreys BD (2018) Mechanisms of Renal Fibrosis. Annu Rev Physiol 80: 309-326. Link: https://bit.ly/2YqIhqo
- Qiao YC, Chen YL, Pan YH, Ling W, Tian F, et al. (2017) Changes of transforming growth factor beta 1 in patients with type 2 diabetes and diabetic nephropathy: A PRISMA-compliant systematic review and meta-analysis. Medicine (Baltimore) 96: e6583. Link: https://bit.ly/3aV7uhN
- Zhou T, Li HY, Zhong H, Zhong Z (2018) Relationship between transforming growth factor-beta1 and type 2 diabetic nephropathy risk in Chinese population. BMC Med Genet 19: 201. Link: https://bit.ly/34qKaaw
- Yang XH, Feng SY, Yu Y, Liang Z (2018) Study on the relationship between the methylation of the MMP-9 gene promoter region and diabetic nephropathy. Endokrynol Pol 69: 269-275. Link: https://bit.ly/3aThh7W
- Garcia-Fernandez N, Jacobs-Cacha C, Mora-Gutierrez JM, Vergara A, Orbe J, et al. (2020) Matrix Metalloproteinases in Diabetic Kidney Disease. J Clin Med 9. Link:
- Zakiyanov O, Kalousova M, Zima T, Tesar V (2019) Matrix Metalloproteinases in Renal Diseases: A Critical Appraisal. Kidney Blood Press Res 44: 298-330. Link: https://bit.ly/3jbnYFk
- Gil-Bernabe P, D'Alessandro-Gabazza CN, Toda M, Boveda Ruiz D, Miyake Y, et al. (2012) Exogenous activated protein C inhibits the progression of diabetic nephropathy. J Thromb Haemost 10: 337-346. Link: https://bit.ly/2EkjhKL
- Selvaraj G, Kaliamurthi S, Kaushik AC, Khan A, Wei YK, et al. (2018) Identification of target gene and prognostic evaluation for lung adenocarcinoma using gene expression meta-analysis, network analysis and neural network algorithms. J Biomed Inform 86: 120-134. Link: https://bit.ly/2EcFHOd
- Cui Y, Liu S, Cui W, Gao D, Zhou W, et al. (2017) Identification of potential biomarkers and therapeutic targets for human IgA nephropathy and hypertensive nephropathy by bioinformatics analysis. Mol Med Rep 16: 3087-3094. Link: https://bit.ly/3gor47l
- HN R, D T, DC C, AM H, F E, A B, A H, CC B, V N, CD C, JW S and M K. Expression data from human with IgA nephropathy. 2012; Link:
- Shved N, Warsow G, Eichinger F, Hoogewijs D, Brandt S, et al. (2017) Transcriptome-based network analysis reveals renal cell type-specific dysregulation of hypoxia-associated transcripts. Sci Rep 7: 8576. Link: https://go.nature.com/32f09ps
- Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, et al. (2011) Transcriptome analysis of human diabetic kidney disease. Diabetes 60: 2354-2369. Link: https://bit.ly/2YqIAS4
- Berthier CC, Bethunaickan R, Gonzalez-Rivera T, Nair V, Ramanujam M, et al. (2012) Cross-species transcriptional network analysis defines shared inflammatory responses in murine and human lupus nephritis. J Immunol 189: 988-1001. Link: https://bit.ly/2FLZZy9
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307-315. Link: https://bit.ly/2YnKByD
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28: 882-883. Link: https://bit.ly/3lfrTTo
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, et al. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. Link: https://bit.ly/3leglQj
- Sun X, Li J (2013) pairheatmap: comparing expression profiles of gene groups in heatmaps. Comput Methods Programs Biomed 112: 599-606. Link: https://bit.ly/2EqYLrJ
- Gene Ontology C (2006) The Gene Ontology (GO) project in 2006. Nucleic Acids Res 34: D322-326. Link: https://bit.ly/32nNnVC
- Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284-287. Link: https://bit.ly/2EoRWXv
- Walter W, Sanchez-Cabo F, Ricote M (2015) GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31: 2912-2914. Link: https://bit.ly/2FSalwE
- Khan A, Mathelier A (2017) Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics 18: 287. Link: https://bit.ly/3aRwc2P
- Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27-30. Link: https://bit.ly/3gpCOqe
- Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, et al. (2010) GeneCards Version 3: the human gene integrator. Database (Oxford) 2010: baq020. Link: https://bit.ly/3l62njw
- Li Y, Li L, Zeng O, Liu JM, Yang J (2017) H2S improves renal fibrosis in STZ-induced diabetic rats by ameliorating TGF-beta1 expression. Ren Fail 39: 265-272. Link: https://bit.ly/2YoZcty
- Zeng M, Liu J, Yang W, Zhang S, Liu F, et al. (2019) Multiple-microarray analysis for identification of hub genes involved in tubulointerstial injury in diabetic nephropathy. J Cell Physiol . Link: https://bit.ly/2FQWKWq
- Sun MY, Wang SJ, Li XQ, Shen YL, Lu JR, et al. (2019) CXCL6 Promotes Renal Interstitial Fibrosis in Diabetic Nephropathy by Activating JAK/STAT3 Signaling Pathway. Front Pharmacol 10: 224. Link: https://bit.ly/31lz1pq
- Wang Y, Zhou J, Minto AW, Hack BK, Alexander JJ, et al. (2006) Altered vitamin D metabolism in type II diabetic mouse glomeruli may provide protection from diabetic nephropathy. Kidney Int 70: 882-891. Link: https://bit.ly/34o9ZYV
- Mou X, Zhou DY, Liu YH, Liu K, Zhou D (2019) Identification of potential therapeutic target genes in mouse mesangial cells associated with diabetic nephropathy using bioinformatics analysis. Exp Ther Med 17: 4617-4627. Link: https://bit.ly/2Ekaclb
- Tervaert TW, Mooyaart AL, Amann K, Cohen AH, Cook HT, et al. (2010) Pathologic classification of diabetic nephropathy. J Am Soc Nephrol 21: 556-563. Link: https://bit.ly/3leh3gr
- Toba H, Lindsey ML (2019) Extracellular matrix roles in cardiorenal fibrosis: Potential therapeutic targets for CVD and CKD in the elderly. Pharmacol Ther 193: 99-120. Link: https://bit.ly/2QjfiR1
- Meng XM, Nikolic-Paterson DJ, Lan HY (2016) TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol 12: 325-338. Link: https://go.nature.com/3ho9ull
- Han Q, Zhu H, Chen X, Liu Z (2017) Non-genetic mechanisms of diabetic nephropathy. Front Med 11: 319-332. Link: https://bit.ly/3aPVoqg
- VR ALBVR, Tan SH, Candasamy M, Bhattamisra SK (2019) Diabetic nephropathy: An update on pathogenesis and drug development. Diabetes Metab Syndr 13: 754-762. Link: https://bit.ly/3gmpbIf
- Flyvbjerg A (2017) The role of the complement system in diabetic nephropathy. Nat Rev Nephrol 13: 311-318. Link: https://bit.ly/2ElvXkr
- Khoshjou F, Dadras F (2014) Mitochondrion and its role in diabetic nephropathy. Iran J Kidney Dis 8: 355-358. Link:
- Darshi M, Van Espen B, Sharma K (2016) Metabolomics in Diabetic Kidney Disease: Unraveling the Biochemistry of a Silent Killer. Am J Nephrol 44: 92-103. Link: https://bit.ly/3aRwRkP
- Hao L, Shan Q, Wei J, Ma F, Sun P (2019) Lactoferrin: Major Physiological Functions and Applications. Curr Protein Pept Sci 20: 139-144. Link: https://bit.ly/31oEP1w
- Brosicke N, Faissner A (2015) Role of tenascins in the ECM of gliomas. Cell Adh Migr 9: 131-140. Link: https://bit.ly/2Yvntyh
- Chiquet-Ehrismann R, Orend G, Chiquet M, Tucker RP, Midwood KS (2014) Tenascins in stem cell niches. Matrix Biol 37: 112-123. Link: https://bit.ly/3lehnvF
- Midwood KS, Valenick LV, Hsia HC, Schwarzbauer JE (2004) Coregulation of fibronectin signaling and matrix contraction by tenascin-C and syndecan-4. Mol Biol Cell 15: 5670-5677. Link: https://bit.ly/34raocR
- Franz M, Jung C, Lauten A, Figulla HR, Berndt A (2015) Tenascin-C in cardiovascular remodeling: potential impact for diagnosis, prognosis estimation and targeted therapy. Cell Adh Migr 9: 90-95. Link: https://bit.ly/2Eqi9oC
- Schwenzer A, Jiang X, Mikuls TR, Payne JB, Sayles HR, et al. (2016) Identification of an immunodominant peptide from citrullinated tenascin-C as a major target for autoantibodies in rheumatoid arthritis. Ann Rheum Dis 75: 1876-1883. Link: https://bit.ly/3aOahJM
- Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, et al. (2011) Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med 17: 867-874. Link: https://bit.ly/3htrwCA
- Fu H, Tian Y, Zhou L, Zhou D, Tan RJ, et al. (2017) Tenascin-C Is a Major Component of the Fibrogenic Niche in Kidney Fibrosis. J Am Soc Nephrol 28: 785-801. Link: https://bit.ly/2Yr4Xa6
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
