Imaging Journal of Clinical and Medical Sciences
Mass Spectrometry Technology for Protein Biomarker Discovery
Sermin Tetik*
Cite this as
Tetik S (2016) Mass Spectrometry Technology for Protein Biomarker Discovery. Imaging J Clin Medical Sci 3(1): 014-016. DOI: 10.17352/2455-8702.000029Review
Human tissues and organs could be affected depending on pathological condition of diseases. Proteins are basic functioning molecules of cells accordingly to modification intensities. Protein modification can be different in cellular interaction, localization, activity, protein concentration, and co-/posttranslational. Protein biomarker discovery has covered some subtitles or points such as differentially expressed proteins disease specific protein isomers and abnormal protein activity [1,2].
Proteomic analysis and conclusion have allowed the potential protein biomarker discovery and thus diagnosis of disease and prognosis can be possible. MS-based proteomic has correlated between protein modification and a certain disease platform to accurately and confidently measure the proteins in serum or plasma, which are low-abundance concentration [3]. MS-based quantitative platform use for tissue samples or several body fluids on protein biomarker discovery. Human fluids such as gastric fluid, urine, and blood have also been evaluated in the last decade. Modifications of proteins as posttranslational can also be indicative of a disease or its progression. Activity-based protein platform changes have allowed for proteome researches of enzymatic and protein-drug interaction events [4,5].
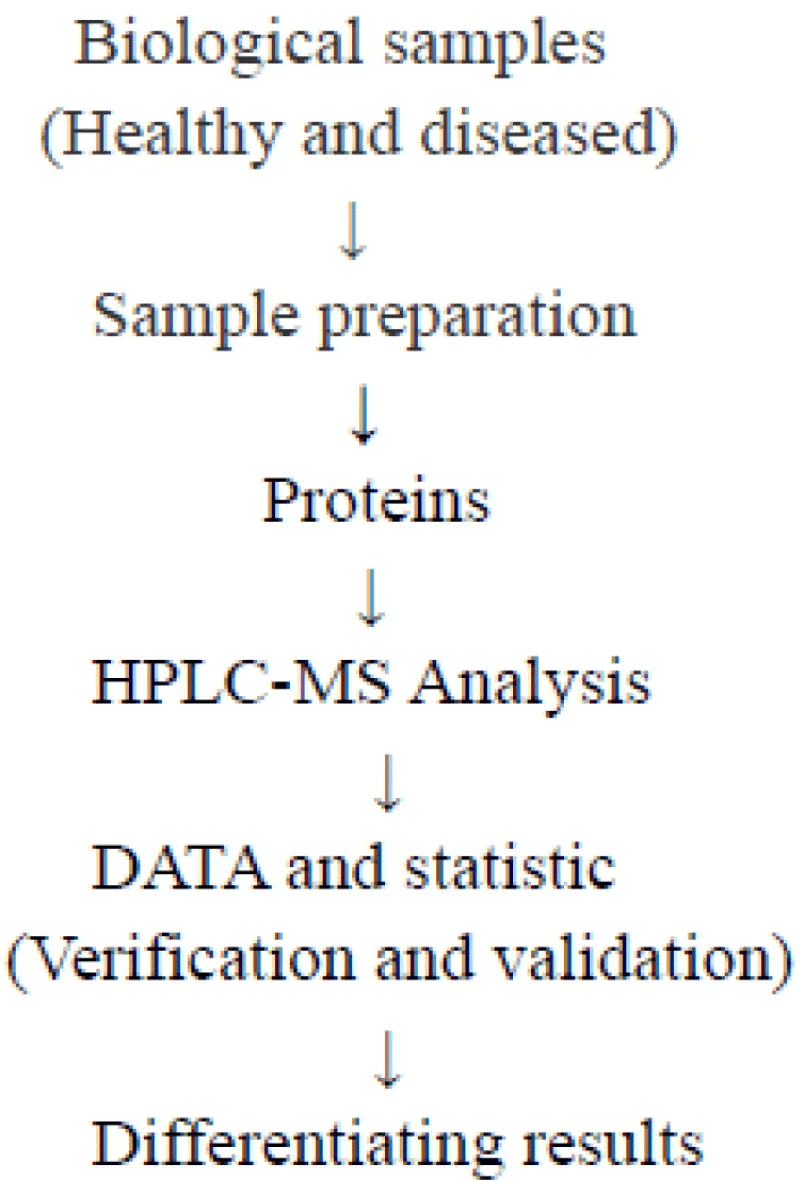
Development of MS-based protein biomarkers is multistep and analogues to drug discovery. The procedure for protein biomarker study has summarized in Figure 1 [6].
Candidate biomarker molecules have different type of genetic sequence such as mRNA transcript, gene variations, SNP variants and all of them can be used as disease biomarkers of protein. If you are choosing proteins as biomarkers, you have an advantage because proteins have diversity. There is approximately 100,000 mRNA transcript and 1,8 million different protein species. Therefore we have to find specific variant of protein in biological samples and measure or detect as correctly. At the biomarker studies pipeline, up to thousand protein species are detected in biological samples. During the biomarker identification stage can choose false positive protein marker depends on the preferred methodological techniques. FDA has approved in vitro diagnostic assay for more than 200 unique proteins, but the majority of them are ELISA-based. And there are more efforts to use mass spectrometry-based protein assay in the clinics [2].
Mass spectrometry-based protein studies have two major types, one is top down and the other is bottom up. Top-down proteomics approaches are used to identify intact protein. Top-down methodology retains especially information about posttranslational modifications of individual residues within proteins, however, clinical applications of top-down proteomics are still limited. Bottom-up proteomics is routinely used for identification at the peptide level and provides peptide fragmentation and sensitivity in biological samples. In analysis of protein biomarker identification, bottom-up proteomics alleviates some advantages such as lower amount of sample. Process of protein identification by bottom-up proteomic methods involves a set of consecutive steps, such as protein digestion, peptide separation by LC, peptide ionization, gas-phase peptide separation, peptide fragmentation, and detection of mass-to-charge ratios (m/z) and intensities of peptide ions and their tandem mass spectrometry (MS/MS) fragments [7,8].
Protease, which is most widely used enzyme, is a trypsin that cleaves of proteins into short peptide fragments C-terminal to lysine and arginine residues. Matrix-assisted laser desorption ionization (MALDI) and ESI use the transfer of large biomolecules into the gas phase without significant structural decomposition. Time-of-flight mass spectrometry (MALDI-TOF) has allow that facilitated analysis of small molecules and intact proteins in cells and human tissues. Imaging mass spectrometry (IMS) provides distribution of intensities of protein, peptide, and small-molecule ions and replaces immunohistochemical-staining methods of tissues and facilitate of tissue biomarkers [9]. ESI is currently the most widely used technique for the mass-based biomarker discovery. In this technique, high voltage application to sample, form of highly charged droplets that eventually evaporate, ions enter the mass spectrometer. If there is high abundance peptide, leads to as ionization suppression to low-abundance peptide. MALDI-MS is used in combination with the time-of-flight (TOF) because TOF instruments have been capable of high mass accuracy; they are used for studies of top-down proteomics and post-translational modifications. Ion-trapping (IT) instruments have high sensitivity, fast scanning speeds, and offer the ability to perform multiple levels of fragmentation of the same substrate. Triple quadrupole mass spectrometers provide increasing selectivity of analysis because filtering of precursor peptide ions and filtering of fragments. The hybrids TOF instruments also provide high mass accuracy (2to 5 ppm) and resolution (10,000 to 40,000), high sensitivity in MS/MS modes, and reduce scan time [4,10].
There is important reducing the number of false-positive peptide spectrum matches by high -resolution-accuracy instruments in mass-based biomarker discovery. Some spectra stem from peptides with posttranslational modifications (PTMs) that are not identification in the search algorithm, from peptides with SNPs, mis-cleaved peptides, solvent ions, contaminants small molecules, lipids, or even airborne molecules of building materials. PTMs of peptides due to pathological conditions are poorly ionized by ESI that does not search for all possible PTMs. To enable efficient PTMs analysis, multiple approaches to enrich PTM peptides can be used. High-resolution mass spectrophotometers provide more potential to enable top-down analysis of PTM in pathological states [11,12].
Identification period of protein biomarkers involve serious technological limitations and biological factors such as intra-individual variations of protein levels during the day in healthy and diseased individuals. Even though hundreds potential biomarkers are selecting, the rate of approved protein biomarkers is decreasing in the last period because that biomarker verification and validation phases involve difficulties of process and high number of false-positive potential biomarkers [13,14].
Proteomic technology is continuing to evolve and improve such as increased automation, sensitivity, miniaturization, and better bioinformatics. On the other hand, limitations of protein biomarker development studies stem from biological factors, such as protein stability or concentrations, pre-analytical parameters. To minimize pre-analytical errors, all samples should be collected, stored, and processed using approved and defined standard procedures.
Basic problems of biomarker discovery and mass spectrometry studies include:
• MS-based measurement is generally relative, should be correlation quantitative relationship between ion intensity and the amount of substrate
• Protein assays are low reproducibility due to day-to-day variability because of multiple steps in bottom-up proteomic process.
There is the methodological limitations of biomarker discovery and mass spectrometry techniques, but new insight of biomarker discovery procedure should improve of approved biomarkers, clinical approaches that prompted biomarker discovery.
- Legrain P, Aebersold R, Archakov A, Bairoch A, Bala K, Beretta L, et al. (2011) The human proteome project: current state and future direction. Mol Cell Proteomics 10: M111.009993. Link: https://goo.gl/fTwXon
- Anderson NL, Anderson NG (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 1: 845-867. Link: https://goo.gl/Xt1SDI
- Li J, Kelm KB, Tezak Z (2011) Regulatory perspective on translating proteomic biomarkers to clinical diagnostics. J Proteomics 74: 2682-2690. Link: https://goo.gl/wUQx2m
- Caprioli RM, Farmer TB, Gile J (1997) Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem 69: 4751-4760. Link: https://goo.gl/KN4rGY
- Tetik S (2016) Proteomics in Cancer Biomarker Discovery. J Pharm Anal Insight 1: Link: https://goo.gl/aPqp2q
- Picotti P, Aebersold R (2012) Selected reaction monitoring- based proteomics: workflows, potential, pitfalls and future directions. Nat Meth 9: 555-566. Link: https://goo.gl/OvvulP
- Kellie JF, Catherman AD, Durbin KR, Tran JC, Tipton JD, et al. (2012) Robust analysis of the yeast proteome under 50 kDa by molecular-mass-based fractionation and top-down mass spectrometry. Anal Chem 84: 209-215. Link: https://goo.gl/hJWj3a
- Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246: 64-71. Link: https://goo.gl/xCmtZS
- Schwamborn K, Caprioli RM (2010) Molecular imaging by mass spectrometry looking beyond classical histology. Nat Rev Cancer 10: 639-646. Link: https://goo.gl/2AKQxq
- Andrews GL, Simons BL, Young JB, Hawkridge AM, Muddiman DC (2011) Performance characteristics of a new hybrid quadrupole time-of-flight tandem mass spectrometer (TripleTOF 5600). Anal Chem 83: 5442-5446. Link: https://goo.gl/Y3fYVK
- Crutchfield CA, Thomas SN, Sokoll LJ, Chan DW (2011) Advances in mass spectrometry-based clinical biomarker discovery. Clin Proteom 13: 1. Link: https://goo.gl/8pmA6D
- Karsdal MA, Henriksen K, Leeming DJ, Woodworth T, Vassiliadis E, et al. (2010) Novel combinations of Post-Translational Modification (PTM) neo-epitopes provide tissue-specific biochemical markers dare they the cause or the consequence of the disease? Clin Biochem 43: 793-804. Link: https://goo.gl/UEe7Vd
- Parker CE, Borchers CH (2014) Mass spectrometry based biomarker discovery, verification, and validation--quality assurance and control of protein biomarker assays. Mol Oncol 8: 840-858. Link: https://goo.gl/G2Quwd
- \Siegfried Neumann (2015) Biomarkers, past and future. Biomarker validation; Technological, clinical and commercial aspects. Eds: herald Seitz, Sarah Schumacher Wiley, VCH. ISBN: 978-3-527-33719-4. Link: https://goo.gl/tvUgJo
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
