Authors:
Elena Kamilari, Yiorgos Apidianakis* and Myrofora Panagi*
Department of Biological Sciences, University of Cyprus, Nicosia, Cyprus
Received: 22 May, 2017; Accepted: 02 June, 2017; Published: 06 June, 2017
Yiorgos Apidianakis, 1 Panepistimiou Ave, Department of Biological Sciences, University of Cyprus, 2109, Nicosia, Cyprus, Tel. 357-22893767; E-mail:
Kamilari E, Apidianakis Y, Panagi M (2017) Flies to Humans - Humans to Flies: A Virtuous Circle of Colorectal Cancer Prevention. Arch Clin Gastroenterol 3(3): 047-060. 10.17352/2455-2283.000038
© 2017 Kamilari E, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Biomarkers; Drosophila; Human; Model host; Colorectal cancer; Inflammation
The two Nobel prizes in physiology or medicine of 1995 and 2011 establish Drosophila genetics as a significant contributor of genes and signaling pathways relevant to human disease, including innate immunity and cancer. Other than providing clues on mammalian gene homologue function, relatively little attention has been paid on the translational aspect of Drosophila genes, microbes and environmental factors that influence homeostasis and disease. This is particularly important for colorectal cancer (CRC) prevention, for which molecular diagnostic tools are non-existent. While clinical studies provide a wealth of information on genes and microbes linked to inflammatory bowel disease (IBD) and CRC, it is unknown if they can serve as biomarkers in terms of CRC prevention. We discuss the line of research showing that many biomarkers of intestinal inflammation and CRC in humans may be modeled and mechanistically tested in flies. Vise versa, genes and processes, such as regenerative inflammation and aging-associated DNA damage, found in flies to promote tumorigenesis may be tested as biomarkers of CRC risk in humans. Thus, modeling human intestinal inflammation and cancer in flies can provide a means to assess causality of conserved genes and microbes that can colonize the fly intestine. Moreover, successful modeling in flies enables the “treatability” of the pertinent biomarkers via dietary, probiotic and pharmacological interventions and paves the way for clinical trials of treatments that may alleviate intestinal inflammation and the risk for CRC.
Introduction
The genetic and histological suitability of flies for basic research on CRC
During the last two decades Drosophila has become a powerful model for exploring the links between inflammation and colorectal cancer (CRC). The fully sequenced genome, the high degree of human disease-related gene homology with flies (up to 75%), the reduced genetic redundancy, the great availability of genetic tools enabling spatial and temporal manipulation of cells, as well as, the evolutionary conservation of signaling pathways controlling vital biological processes and immunity, make Drosophila a suitable model host for the identification of candidate biomarkers implicated in tumor-promoting inflammation [1,2]. In addition, the small size, the low cost of maintenance and the easy delivery of orally administrated drugs facilitate whole-animal screening for molecular compounds affecting stem cell mediated carcinogenesis [3,4]. Remarkably, there are at least 60 chemical compounds originally known for their activity in human cells that demonstrably have the same molecular mechanism of action in flies [2].
The intestine is the most rapidly self-renewing tissue of the human body. Intestinal epithelium is continuously exposed to pathogens and chemicals of the lumen leading to enterocyte damage and concomitant regenerative inflammation that completely replenishes the damaged or lost cells by asymmetrically dividing intestinal stem cells (ISCs) [5-7]. ISCs reside at the bottom of the crypt of Lieberkühn along with the secretory Paneth cells. ISCs proliferate to give rise to transient cells that amplify and differentiate, while moving upwards the crypt. Differentiated cells are found at the rim of the crypts and the villi. These are absorptive enterocytes, but also secretory cells, namely, Paneth, enderoendocrine or goblet cells. Between the villi and the lumen there is a goblet cell derived-mucus layer that protects cells from direct bacterial contact. The tissue is supported by stromal cells of various types and surrounded by visceral muscle. Similarly, Drosophila intestine is maintained by ISCs that divide giving rise to new ISCs and transient enteroblasts, which normally differentiate without further divisions into either absorptive enterocytes or enteroendocrine cells [8,9]. Although the mammalian Paneth, goblet and stromal cells are absent in flies, many of their immunity and barrier physiology functions are fulfilled by highly endoreplicating enterocytes and the visceral muscle [1]. The fly intestine generally lacks crypts that support multiple progenitor cells and villi. These appear necessary for maximal nutrient absorption in mammals, but in flies a monolayer of epithelial cells surrounded by two layers of visceral muscle suffices for homeostasis. In addition to the mucus layer, the Drosophila gut lumen is surrounded by a chitin layer, the peritrophic matrix that confers extra protection against pathogens [10].
Using flies for identifying genetic and microbial biomarkers of risk for CRC
Fly immunity shares evolutionary conserved mechanisms with human innate immunity. Drosophila midgut responds to uracil from intestinal pathogenic bacteria by inducing the p38 mitogen-activated protein kinase (p38 MAPK) signaling pathway [11,12], which in turn activates the conserved NAPDH oxidase, Duox, leading to the release of reactive oxygen species (ROS) [13-15]. The antimicrobial activity of ROS is complemented with the production of antimicrobial peptides (AMPs). Toll signaling pathway is responsible for the systemic AMP response mediated by the nuclear translocation of the NF-κB-like transcription factor(s) Dorsal and/or Dif. The second NF-κB-like pathway of Drosophila, named immune deficiency (IMD) pathway, is stimulated by peptidoglycan recognition proteins (PGRPs). IMD is induced by bacterial peptidoglycan leading to the nuclear translocation of Relish and AMPs expression both systemically and in the fly gut [16,17]. Intestinal damage and stress are also capable of stimulating particular AMP expression following secretion of the Upd3 inflammatory cytokine, an analog of the human interleukin (IL)-6, which activates the JAK/STAT signaling in both ISCs and the visceral muscle [18,19]. JAK/STAT pathway activation leads to the expression of epidermal growth factors and consequently induction of EGFR/Ras/MAPK cascade inducing ISC proliferation [20,21]. Drosophila stem cells are further modulated by the Target of Rapamycin (TOR), Hippo and wingless pathways [22-24]. This inflammation induced tissue regeneration process referred to as regenerative inflammation may contribute to tumor initiation and progression and is conserved between flies and mammals [6,7].
Inflammation is pivotal for host defense, but it can lead to pathogenesis when chronic and predispose for cancer. The inflammatory microenvironment facilitates tumor initiation and progression, although a direct causality has not been established for CRC. Germline mutations account as a driving force for the 10% of CRC incidence, whereas the vast majority of cases associate with somatic mutations and environmental factors, including chronic inflammation and bacterial infections [25,26]. For instance, the chronic inflammation of the gastrointestinal mucosa present in IBD patients is a key predisposing factor for developing CRC [27]. Nevertheless, so far only Helicobacter pylori infection has been established as a causative agent of gastrointestinal inflammation and cancer [28]. In mammals, areas with active inflammatory responses accompanied by a high rate of epithelial cell-turnover and sustained DNA damage are sufficient to drive carcinogenesis [29]. This setting of increased predisposition to tumorigenesis is also found in the aging midgut of Drosophila [30]. Moreover, pathogenic bacterial infection promotes intestinal tumorigenesis in genetically predisposed adult Drosophila [31].
Previous studies have used fruit flies as model hosts to induce intrinsic and extrinsic oxidative damage that resembles the aging associated changes in progenitor cells [32]. Collective evidence indicates accumulation of γH2AvD foci in ISCs, analogous to the mammalian γH2AX, a DNA damage marker. γH2AvD foci correlate with γ-ray-induced DNA damage in ISCs and age-related accumulation of 8-oxo-2′deoxyguanosine. Interestingly, age and oxidative stress related DNA damage in Drosophila can be alleviated by the chemotherapy drug Metformin via downregulation of the insulin-like growth factor-I receptor/insulin receptor (IGF1R/IR) and the AKT [33]. As ISCs age, they acquire persistent chromatin lesions bearing double strand breaks (DSB) and thereby, initiating a continuous secretion of inflammatory cytokines. In mammals, damaged ISCs that have entered senescence preserve their capacity to secrete different factors and interact with the surrounding microenvironment [34]. Similarly, age-related DNA damage and JNK-driven dysplasia correlate with barrier failure and excessive systemic inflammatory signaling attributed to the bacterial translocation across the Drosophila gut [35,36]. Somatic inactivation of Notch tumor suppressor during aging in flies causes spontaneous neoplasia driven by somatic recombination, genomic deletions and rearrangements [30].
As microbial intestinal load increases with age, it becomes challenging to maintain symbiosis between the host and its microbes. Alterations within the microbiome structure could elicit an acute inflammatory signaling through the increased production of ROS and AMPs and the release of inflammatory cytokines and growth factors that regenerate the intestinal epithelium. However, chronic inflammatory responses and the excessive exposure of cells to oxidative stress have adverse effects on homeostasis, leading to dysbiosis. Dysbiosis is correlated with IBD [37] and cancer [38]. Drosophila is characterized by a simple microbiome of less than 30 microbial species [35] compared to that of humans, which is composed with hundreds of different bacterial species [39]. Lactobacillus and occasionally Enterobacteriaceae are part of both the fly and human microbiome. While huge differences exist, symbiotic bacteria are critical for host physiology in both species and the number of human bacteria that can colonize flies is far more than those found naturally. They promote growth by modulating nutrient metabolism and absorption [40] and participate in the shaping of gastrointestinal immune landscape [41,42]. Therefore, pinpointing the mechanisms by which gut microbiota affects health and disease, may help to suggest new therapeutic approaches to alleviate microbiota-directed inflammation and CRC incidence. Bacterial mono-associations or poly-associations with germ-free flies would provide insights regarding the contribution of symbiotic bacteria at the species level in intestinal disease.
Mammalian genetic and microbial biomarkers of risk for CRC
Most CRC cases are sporadic, that is, with no known genetic component attributed to them (70%-80% of all cases) and usually appear at an old age [43,44]. Hereditary forms of CRC include familial adenomatous polyposis (~1%), non-polyposis hereditary CRC or Lynch syndrome (2%-5%) and MYH-gene associated polyposis (<1%) [45]. Interestingly, molecular and cellular alterations precede morphological changes of the intestinal mucosa, and may predispose for tumorigenesis [46,47]. This must be true also for intestinal microbes and their balance [48]. Such alterations may be blamed for the recurrence of adenomatous polyps after surgical excision [49]. Therefore, ongoing efforts turn towards finding specific genetic and microbial markers that will allow the early detection of CRC appearance in terms of personalized medicine and treatment.
A hallmark of transition from normal colonic epithelium to neoplastic is genomic instability (GI), which is divided to chromosomal instability (CIN), microsatellite instability (MSI) and epigenetic instability (EI) [44]. The most familiar form of GI is CIN, which is implicated in 80%–85% of colorectal tumors [50]. GI is characterized by a) the presence of aneuploidy, which involves changes in chromosome number, b) modifications in the gene structure, such as insertions, deletions or base substitutions, which are also caused by MSI, c) chromosome rearrangements and d) gene multiplying. The basic concept of GI involves the loss of function of tumor-suppressor genes, such as adenomatous polyposis coli (APC), p53, SMAD4, and tumor-suppressor genes on chromosome 18q the area deleted in colon cancer (DCC), or the activation of the ,K-Ras oncogene [51].
APC is mutated in up to 80% of sporadic CRC cases and is involved in the negative regulation of Wingless-Int (WNT) signaling pathway. WNT pathway regulates various biological processes, such as cell proliferation, differentiation, polarity, and movement, and maintains intestinal epithelial cell (IEC) homeostasis [52-54]. The canonical WNT pathway is highly conserved and initiates with the binding of Wnts to Frizzled (Fz) receptors [55-57]. Downstream of Fz, Dishevelled (Dsh) the glycogen synthase kinase 3 beta (GSK3β) and casein kinase I-γ (CK1γ) result in the docking of the scaffold protein Axin and APC and the stabilization of β-catenin. The complex with β-catenin is disrupted upon ligand signaling and β-catenin moves to the nucleus where it binds to T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors activating specific Wnt target genes. In the absence of Wnt, the serine/threonine kinases, CK1 and GSK3α/β, phosphorylate β-catenin, which is guided by the F box/WD repeat protein β-TrCP, for degradation to the proteasome. In the absence of signaling, TCF/LEF is not activated by β-catenin and its targets are suppressed by Groucho [58]. The TCF binding sites are also similar between vertebrates and Drosophila [59]. The function of β-catenin as a TCF activator is strictly regulated by the multiprotein complex that involves APC, which when mutated β-catenin is activated to induce tumor-promoting genes, such as Myc [60].
The KRAS oncogene is an activated form of the endogenous gene and present in up to 43% of human CRC tumors. It encodes the guanosine diphosphate (GDP) and guanosine triphosphate (GTP) binding proteins. Wild type KRAS is induced by the epidermal growth factor receptor (EGFR) pathway, but the KRAS oncogene is constitutively active independently of such stimulation [61]. Activation of EGFR promotes an excessive mitogenic signaling cascade through the activation of numerous pathways, including the RAS – RAF – mitogen-activated protein kinase (MAPK), the phosphatidylinositol 3-kinase (PI3K) – Akt, and the phospholipase C pathway [62, 63]. BRAF V600E (involved in 10-15% of CRC tumors), which encodes a guanosine triphosphate (GTPase), is also involved in the EGFR pathway activation [64]. PIK3CA gene of PI3K pathway is mutated in ∼15% of CRCs [65], while some cases include mutations in PTEN, a tumor suppressor, which normally inhibits PI3K [66]. Additional mutations present in CRC tumors include TCF7L2, , NRAS, FAM123B, CTNNB1 and SMAD2 [67] (Table 1). Mutations in the MSI pathway are primarily due to the loss of function of DNA repair proteins, MLH1, MLH3, PMS1, PMS2, MSH2, MSH3, MSH6 and Exo1 [68]. MSI also affects cell mitosis (TGF-β, GRB1, TCF-4, WISP3, activin receptor-2, IGF-2 receptor, axin-2, and CDX), apoptosis (BAX, caspase-5, RIZ, BCL-10, PTEN, hG4-1, and FAS), and additional DNA repair genes (MBD-4, BLM, CHK1 and RAD50) [69, 70]. The most known, clinically evaluated MSI markers are mononucleotide (BAT-25, BAT-26, NR-21, NR-24 and MONO-27) exhibiting high sensitivity and specificity [64] (Table 2). Two kinds of EI have been mostly described in CRC: CpG island methylator phenotype (CIMP), and global DNA hypomethylation. Both mechanisms cause silencing of gene expression. Known biomarkers for CIMP-positive tumors are CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1 and methylation must occur in at least 3 of them [71]. Indicative lists of mouse and fly homologs of human genetic and epigenetic biomarkers of inflammation and CRC are provided in Tables 1, 2.
-
Table 2:
Processes and genes affecting susceptibility to intestinal infection, stress or inflammation in humans, mice and flies, per Vaiserman, 2015 [89].
Genetic biomarkers linked to inflammation (IBD) may also be linked to CRC. Chronic inflammation and tissue damage induce cell proliferation and aberrant differentiation of macroscopically normal-appearing colonic mucosa, which may lead to crypt enlargement and potentially to cancer initiation and progression [72-74]. The most used marker for cell proliferation during inflammation and cancer is KI67 [75-80]. Additional markers used to estimate colorectal tumor cell mitosis are MCM7 and its negative regulator Geminin, which are involved in the DNA replication [81-83], as well as, Aurora kinase A (AURKA), which plays a critical role in cell cycle regulation [84], and proliferating cell nuclear antigen (PCNA), which is necessary for DNA synthesis during replication [85,86]. Inflammation responses involve the recruitment of tissue-resident macrophages and mast cells, which produce a variety of inflammatory mediators, including cytokines, chemokines, proteases, matrix metalloproteinases, TNF-α, interleukins (IL), interferons (IFN), and enzymes such as cyclooxygenase-2 (COX-2), 5-lipoxygenase (5LOX), and phospholipase A2 (PLA2) responsible for eicosanoid formation [87]. Many other cytokines may be pro-tumorigenic, including IL-4, IL-6, IL-8, IL-11, IL-17A, IL-22, IL-23, IL-33, TNF, TGF-β, and VEGF [88]. These mediators could serve as prognostic biomarkers for CRC appearance. Additional processes and genes affecting susceptibility to intestinal infection, stress or inflammation in human, mice and flies are described in table 2 [89].
Bacteria have been linked both positively and negatively to inflammation (IBD) and CRC. Major disruption of the healthy microbiota caused by extensive and prolonged antibiotic use, especially in neonates and children, can result in life-threatening necrotizing enterocolitis. In this case intestinal dysbiosis results in excessive inflammation, mucosal injury and cell death without regeneration [90]. Shifting the balance of intestinal microbiota from a pathogenic to a protective complement of bacteria can protect the gut from inflammation and subsequent injury [91]. Accordingly, broad-spectrum antibiotics destroy the flora leading to intestinal inflammation and damage that can be prevented with oral administration of microbiota-derived molecular patterns, such as lipopolysaccharide (LPS) that induce a steady state of inflammatory cytokines and prime the epithelium again pathogens [92]. From the immunological perspective evidence reveals various properties of intestinal bacteria that distinguish them as commensals vs. pathogens [93]. Nevertheless, for the most part microbiota is just linked to the healthy or the diseased state with only a few clear cases of bacterial pathogens presented as causal for the disease. Reduced abundance of potentially beneficial bacteria has been reported for patients with IBD and CRC (Table 3). Some of these belong to the family Lachnospiraceae, which produce short chain fatty acids, such as the anti-inflammatory butyric acids [94] and include the genera Lachnospira [95-97], Blautia [95,98-102], Anaerostipes [95] and Roseburia [103-106], even though some reports indicate increase abundance in CRC [107,108]. The genus Lactobacillus was negatively correlated with CRC [109-115]. Lactobacillus strains have been well characterized for their probiotic properties including production of butyrate, which has an antiinflammatory function [94], as well as, the antibacterial lactic acid and bacteriocins. In addition, Lactobacilli reduce the secretion of virulence factors from enterovirulent pathogens alleviating their deleterious effects on the host [116]. Similarly, Clostridia members of clusters XIVa, IV and XVIII, have been reported to reduce inflammation [117] and have decreased abundance in IBD [117-122]. Moreover, some reports have found reduced amounts of Bifidobacteria in CRC patients [95], while others the opposite [123].
-

Table 3:
Genera over/under represented in IBD and CRC and their oxygen sensitivity.
Translational studies for identifying “treatable” biomarkers of risk for CRC using Drosophila
Deciphering the right inflammatory status in the intestine is necessary for designing clinical trials against IBD and maybe CRC. Mucosal healing in IBD patients has shown promise as it correlates with remission of ulcers in Crohn’s disease and erosions and ulcers in ulcerative colitis [7,124]. Transcriptomic studies in humans have led to the identification of genetic and microbial associations with IBD and CRC. Here, we emphasize the use of Drosophila as a whole-animal model to validate the effectiveness, causality and toxicity of identified “treatable” biomarkers in intestinal disease. Examples include the “humanized” Drosophila strains, which are genetically engineered to express human orthologs [2,125]. Notably, tens of chemicals originally selected to target human proteins, such as Rapamycin, BEZ235, SP600125 and DAPT, have been shown to have the same mechanism of action in Drosophila i.e. inhibition of PI3K/mTOR, JNK and γ-secretase/Notch signaling, respectively [125-130]. Thus, accumulating evidence suggests that Drosophila could fill the gap between in vitro and mammalian model host testing (Figure 1).
-
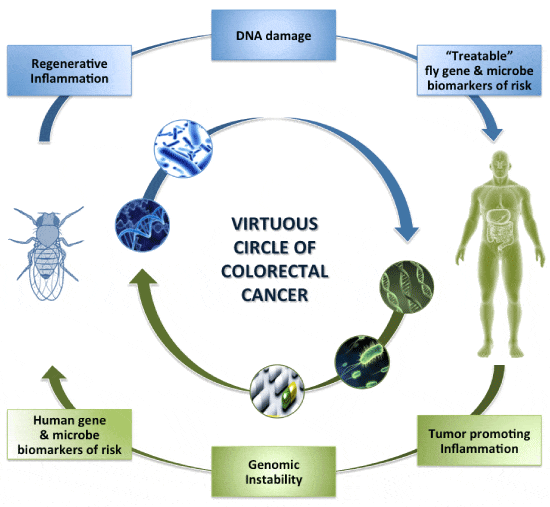
Figure 1:
A virtuous circle of research on CRC prevention. Work on the facilitating hallmarks of cancer, tumor-promoting inflammation and genomic instability, can in addition to unbiased approaches identify genetic and microbial biomarkers linked to risk for CRC. Biomarkers can be further tested and manipulated therapeutically by diet, drug and probiotic administration in flies to assess causality and facilitate clinical trials in humans.
Moreover, flies could be used to examine the association between identified intestinal disease-related microbiota and host. For instance, the commensal microbe of Drosophila Acetobacter pomorum was found to modulate insulin pathway via acetic acid production and subsequently promote ISC proliferation and overall animal growth [131]. Also, a positive correlation between Enterococcus spp. and IBD patients has been also reported. Enterococcus strains that form better biofilms, adhere strongly on intestinal cells and possess antioxidant defense mechanisms are mostly found in IBD patients versus healthy people [132]. Therefore, dissecting host-microbiome interactions of overrepresented in IBD and CRC bacterial strains, such as Bacteroides, Escherichia, Enterococcus and Enterobacter in gnotobiotic flies could give insights regarding bacterial pathogenicity. This is more feasible nowadays due to the increasing number of human microbiota species that we are able to culture and thus test in model organisms, such as flies [133]. Similarly, Drosophila could be used to determine the beneficial impact of bacteria underrepresented in CRC like Clostridia and Lactobacillus.
Accumulated evidence in Drosophila highlights the role of diet in intestinal disease. Nutrient deprivation and reduced insulin pathway correlate with reduced ISC proliferation and number, a phenotype that is reversible upon feeding [134]. Dietary L-glutamate also stimulates intestinal cell proliferation and growth via regulation of Ca2+ signaling [135]. This plasticity of ISC to nutrient availability could be used to target the aberrant proliferation of dysplastic lesions. Given that CRC is a multifactorial disease, a sophisticated combination of probiotic, chemical and dietary interventions might be required to efficiently prevent the disease. In this regard, tumor-initiating inflammation may be successfully targeted by sequestration of regenerative chemokines/cytokines and selective inhibition of signaling molecules that promote tumor survival and growth [25].
Limitations
The use of animal models provides the ability to study the effects of biomarkers of fundamental signaling pathways, microbes and environmental factors and suggest therapeutic interventions against intestinal inflammation and tumorigenesis. A practical limitation of using Drosophila in translational studies on IBD and CRC is the inability to assess the disease promoting properties of human anaerobes that are highly sensitive to the presence of oxygen. An additional limitation of the fly model is the lack of adaptive immunity and the absence of lamina propria in which immune cells reside and infiltrate. Thus, alternative animal hosts such as mouse models should be used to validate and complement the assessment of biomarkers, especially those related to adaptive immunity and highly sensitive to oxygen microbes. Regardless, advantages such as the short lifespan of the fly facilitate assessments of drug-diet-microbial interventions against sporadic intestinal cancer during ageing that is impractical to perform in mice. Thus, Drosophila can be an attractive model host for studying well-conserved genetic, microbial, and environmental components of intestinal homeostasis and disease, the analogous features of which might play a pivotal role in human health.
- Apidianakis Y, Rahme LG (2011) Drosophila melanogaster as a model for human intestinal infection and pathology. Dis Model Mech 4: 21-30. Link: https://goo.gl/RtfHyD
- Fernandez-Hernandez I, Scheenaard E, Pollarolo G, Gonzalez C (2016) The translational relevance of Drosophila in drug discovery. EMBO reports 17: 471-472. Link:© https://goo.gl/iyeYS3
- Tzelepis I, Kapsetaki SE, Panayidou S, Apidianakis Y (2013) Drosophila melanogaster: a first step and a stepping-stone to anti-infectives. Curr.Opin.Pharmacol 13: 763-768. Link: https://goo.gl/00Vpv6
- Markstein M, Dettorre S, Cho J, Neumuller RA, Craig-Muller S, et al. (2014) Systematic screen of chemotherapeutics in Drosophila stem cell tumors. Proc Natl Acad Sci U S A 111: 4530-4535. Link: https://goo.gl/OswZmS
- Crosnier C, Stamataki D, Lewis J (2006) Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet 7: 349-359. Link: https://goo.gl/prKGyB
- Panayidou S, Apidianakis Y (2013) Regenerative inflammation: lessons from Drosophila intestinal epithelium in health and disease. Pathogens 2: 209-231. Link: https://goo.gl/epg8kJ
- Karin M, Clevers H (2016) Reparative inflammation takes charge of tissue regeneration. Nature 529: 307-315. Link: https://goo.gl/oRM2Ud
- Micchelli CA, Perrimon N (2006) Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439: 475-479. Link: https://goo.gl/E7y6IN
- Ohlstein B, Spradling A (2006) The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439: 470-474. Link: https://goo.gl/XchMqC
- Kuraishi T, Binggeli O, Opota O, Buchon N, Lemaitre B, et al. (2011) Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc Natl Acad Sci U S A 108: 15966-15971. Link: https://goo.gl/hnsOLM
- Ha E, Lee K, Seo YY, Kim S, Lim J, et al. (2009) Coordination of multiple dual oxidase– regulatory pathways in responses to commensal and infectious microbes in Drosophila gut. Nat Immunol 10: 949-957. Link: https://goo.gl/s6bhcN
- Lee K, Kim S, Kim E, Ha E, You H, et al. (2013) Bacterial-Derived Uracil as a Modulator of Mucosal Immunity and Gut-Microbe Homeostasis in Drosophila. Cell 153: 797-811. Link:© https://goo.gl/uCTdgA
- Ha EM, Oh CT, Bae YS, Lee WJ (2005) A direct role for dual oxidase in Drosophila gut immunity. Science (New York) 310: 847-850. Link: https://goo.gl/dkWD1h
- Bae YS, Choi MK, Lee W (2010) Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends in immunology 31: 278-287. Link: https://goo.gl/amVj7I
- Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, et al. (2013) Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J 32: 3017-3028. Link: https://goo.gl/sPB9qK
- Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart J, et al. (2000) Tissue-Specific Inducible Expression of Antimicrobial Peptide Genes in Drosophila Surface Epithelia. Immunity 13: 737-748. Link: https://goo.gl/lJFCRw
- Buchon N, Osman D, David FA, Yu Fang H, Boquete J, et al. (2003) Morphological and Molecular Characterization of Adult Midgut Compartmentalization in Drosophila. Cell Rep 3: 1725-1738. Link: https://goo.gl/Y5TWBO
- Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B (2009) Drosophila Intestinal Response to Bacterial Infection: Activation of Host Defense and Stem Cell Proliferation. Cell Host Microbe 5: 200-211. Link: https://goo.gl/kl2mDs
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, et al. (2009) Cytokine/Jak/Stat Signaling Mediates Regeneration and Homeostasis in the Drosophila Midgut. Cell 137: 1343-1355. Link: https://goo.gl/pGc83S
- Buchon N, Broderick NA, Kuraishi T, Lemaitre B (2010) Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC biol 8: 152. Link: https://goo.gl/zEBMQu
- Jiang H, Edgar BA (2011) Intestinal stem cells in the adult Drosophila midgut. Exp Cell Res 317: 2780-2788. Link: https://goo.gl/NiUsjC
- Lin G, Xu N, Xi R (2008) Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature 455: 1119-1123. Link: https://goo.gl/eaNRt0
- Ren F, Wang B, Yue T, Yun EY, Ip YT, et al. (2010) Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc Natl Acad Sci USA 107: 21064-21069. Link: https://goo.gl/l8CVxm
- Quan Z, Sun P, Lin G, Xi R (2013) TSC1/2 regulates intestinal stem cell maintenance and lineage differentiation through Rheb-TORC1-S6K but independently of nutritional status or Notch regulation. J Cell Sci 126: 3884-3892. Link: https://goo.gl/e4qN8W
- Grivennikov SI, Greten FR, Karin M (2010) Immunity, inflammation, and cancer. Cell 140: 883-899. Link: https://goo.gl/ERnjzD
- Yurgelun MB, Kulke MH, Fuchs CS, Allen BA, Uno H, et al. (2017) Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol 35: 1086-1095. Link: https://goo.gl/H8mS9e
- Itzkowitz SH, Yio X (2004) Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol. Gastrointestinal and liver physiology 287: G7-17. Link: https://goo.gl/xxOh9D
- de Martel C, Franceschi S (2009) Infections and cancer: established associations and new hypotheses. Crit Rev Oncol Hematol 70: 183-194. Link: https://goo.gl/ZSB4K7
- Coussens LM, Werb Z (2002) Inflammation and cancer. Nature 420: 860-867. Link: https://goo.gl/CdguZz
- Siudeja K, Nassari S, Gervais L, Skorski P, Lameiras S, et al. (2015) Frequent somatic mutation in adult intestinal stem cells drives neoplasia and genetic mosaicism during aging. Cell stem cell 17: 663-674. Link: https://goo.gl/CmlHiu
- Apidianakis Y, Pitsouli C, Perrimon N, Rahme L (2009) Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc Natl Acad Sci USA 106: 20883-20888. Link: https://goo.gl/QkhbHw
- Park J, Lee S, Na H, Pyo J, Kim Y, et al. (2012) Age-and oxidative stress-induced DNA damage in Drosophila intestinal stem cells as marked by Gamma-H2AX. Exp Gerontol 47: 401405. Link: https://goo.gl/cC5z8h
- Na H, Park J, Pyo J, Lee S, Jeon H, et al. (2013) Mechanism of metformin: Inhibition of DNA damage and proliferative activity in Drosophila midgut stem cell. Mech Ageing Dev 134: 381-390. Link: https://goo.gl/2q3obh
- Rodier F, Coppé J, Patil CK, Hoeijmakers WA, Muñoz DP, et al. (2009) Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nature cell biol 11: 973-979. Link: https://goo.gl/uF8KBe
- Buchon N, Silverman N, Cherry S (2014) Immunity in Drosophila melanogaster - from microbial recognition to whole-organism physiology. Nature reviews immunology 14: 796-810. Link: https://goo.gl/Z9ax2f
- Biteau B, Hochmuth CE, Jasper H (2008) JNK Activity in Somatic Stem Cells Causes Loss of Tissue Homeostasis in the Aging Drosophila Gut. Cell stem cell 3: 442-455. Link: https://goo.gl/oYGlBq
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, et al. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007; 104: 13780-13785. Link: https://goo.gl/1PNJJD
- Lupton JR (2004) Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr 134: 479-482. Link: https://goo.gl/oAXtB1
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489: 220-230. Link: https://goo.gl/YPJ44L
- Sommer F, Bäckhed F (2013) The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 11: 227-238. Link: https://goo.gl/7VCA0j
- Lievin-Le Moal V, Servin AL (2006) The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev 19: 315-337. Link: https://goo.gl/di8TRR
- Hooper LV, Littman DR, Macpherson AJ (2012) Interactions between the microbiota and the immune system. Science 336: 1268-1273. Link: https://goo.gl/Nhfd4u
- Whiffin N, Hosking FJ, Farrington SM, Palles C, Dobbins SE, et al. (2014) Identification of susceptibility loci for colorectal cancer in a genome-wide metaanalysis. Hum Mol Genet 23: 4729-4737. Link: https://goo.gl/R99fB6
- Binefa G, Rodríguez-Moranta F, Teule À, Medina-Hayas M (2014) Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol 20: 6786-6808. Link: https://goo.gl/Yktk09
- Farrington SM, Tenesa A, Barnetson R, Wiltshire A, Prendergast J, et al. (2005) Germline susceptibility to colorectal cancer due to base-excision repair gene defects. Am J Hum Genet 77: 112-119. Link: https://goo.gl/cyFW2G
- Nambiar PR, Gupta RR, Misra V (2010) An “Omics” based survey of human colon cancer. Mutat Res 693: 3-18. Link: https://goo.gl/xBxE3L
- Aghagolzadeh P, Radpour R (2016) New trends in molecular and cellular biomarker discovery for colorectal cancer. World J Gastroenterol 22: 5678. https://goo.gl/3lvAYf
- Sears CL, Garrett WS (2014) Microbes, microbiota, and colon cancer. Cell Host Microbe 15: 317-328. Link: https://goo.gl/0xdPGX
- Rutter MD, Chattree A, Barbour JA, Thomas-Gibson S, Bhandari P, et al. (2015) British Society of Gastroenterology/Association of Coloproctologists of Great Britain and Ireland guidelines for the management of large non-pedunculated colorectal polyps. Gut 64: 1847-1873. Link: https://goo.gl/numkFN
- Grady WM, Carethers JM (2008) Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 135: 1079-1099. Link: https://goo.gl/LT9Na6
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, et al. (1988) Genetic alterations during colorectal-tumor development. New England Journal of Medicine 319: 525-532. Link: https://goo.gl/OhdaT9
- Fearnhead NS, Britton MP, Bodmer WF (2001) The ABC of APC. Hum mol genet 10: 721733. Link: https://goo.gl/u1dyfT
- Nusslein-Volhard C, Wieschaus E (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287: 795-801. Link: https://goo.gl/WlsxEu
- Rijsewijk F, Schuermann M, Wagenaar E, Parren P, Weigel D, et al. (1987) The Drosophila homology of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell 50: 649-657. Link: https://goo.gl/NLprsQ
- Bhanot P, Brink M, Samos CH, Hsieh J (1986) A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature 382: 225-230. Link: https://goo.gl/9xjDYQ
- Wehrli M, Dougan ST, Caldwell K, O'keefe L, Schwartz S, et al. (2000) arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature 407: 527. Link: https://goo.gl/oiFc1M
- Culi J, Mann RS (2003) Boca, an endoplasmic reticulum protein required for wingless signaling and trafficking of LDL receptor family members in Drosophila. Cell112: 343-354. Link: https://www.ncbi.nlm.nih.gov/pubmed/12581524
- Clevers H (2006) Wnt/β-catenin signaling in development and disease. Cell 127: 469-480. Link: https://goo.gl/1CWzdB
- Van de Wetering M, Cavallo R, Dooijes D, Van Beest M, Van Es J, et al. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88: 789-799. Link: https://goo.gl/klvy8s
- Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, et al. (2014) Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511: 483-487. Link: https://goo.gl/KRWUjF
- Siddiqui AD, Piperdi B (2010) KRAS mutation in colon cancer: a marker of resistance to EGFR-I therapy. Ann surg oncol 17: 1168-1176. Link: https://goo.gl/jXfzLR
- Yarden Y, Sliwkowski MX (2001) Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2: 127-137. Link: https://goo.gl/6ZxVFd
- Scaltriti M, Baselga J (2006) The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res 12: 5268-5272. Link: https://goo.gl/B5rR6j
- Pritchard CC, Grady WM (2011) Colorectal cancer molecular biology moves into clinical practice. Gut 60: 116-129. Link: https://goo.gl/kBWZPn
- Rosty C, Young JP, Walsh MD, Clendenning M, Sanderson K, et al. (2013) PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival. PloS one 8: e65479. Link: https://goo.gl/1uDPff
- Parsons B, Foley E (2013) The Drosophila platelet-derived growth factor and vascular endothelial growth factor-receptor related (Pvr) protein ligands Pvf2 and Pvf3 control hemocyte viability and invasive migration. J Biol Chem 288: 20173-20183. Link: https://goo.gl/7YTYPq
- Carethers JM (2014) DNA testing and molecular screening for colon cancer. Clin Gastroenterol Hepatol 12: 377-381. Link: https://goo.gl/NMS7zh
- Boland CR, Goel A (2010) Microsatellite instability in colorectal cancer. Gastroenterology 138: 20732087. e3. Link: https://goo.gl/7U149Z
- Markowitz S, Wang J, Myeroff L, Parsons R (1995) Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science 268: 1336-1338. Link: https://goo.gl/onW5kv
- Duval A, Hamelin R (2002) Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer res 62: 2447-2454. Link: https://goo.gl/j7aBQz
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, et al. (2006) CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38: 787-793. Link: https://goo.gl/1UaCyQ
- Bostick RM, Fosdick L, Grandits GA, Lillemoe TJ, Wood JR, et al. (1997) Colorectal epithelial cell proliferative kinetics and risk factors for colon cancer in sporadic adenoma patients. Cancer Epidemiol Biomarkers Prev 6: 1011-1019. Link: https://goo.gl/O7pVmh
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, et al. (2007) Identification and expansion of human colon-cancer-initiating cells. Nature 445: 111-115. Link: https://goo.gl/13QxwY
- Ben-Neriah Y, Karin M (2011) Inflammation meets cancer, with NF-[kappa] B as the matchmaker. Nat immunol 12: 715-723. Link: https://goo.gl/thTbl9
- Porschen R, Lohe B, Hengels K, Borchard F (1989) Assessment of cell proliferation in colorectal carcinomas using the monoclonal antibody KI-67. Correlation with pathohistologic criteria and influence of irradiation. Cancer 64: 2501-2505. Link: https://goo.gl/R2Jspc
- Palmqvist R, Sellberg P, Oberg A, Tavelin B, Rutegard JN, et al. (1999) Low tumour cell proliferation at the invasive margin is associated with a poor prognosis in Dukes' stage B colorectal cancers. Br J Cancer 79: 577-581. Link: https://goo.gl/YYk7pS
- Oshima CT, Iriya K, Forones NM (2005) Ki-67 as a prognostic marker in colorectal cancer but not in gastric cancer. Neoplasma 52: 420-424. Link: https://goo.gl/TpZB3B
- Salminen E, Palmu S, Vahlberg T, Roberts PJ, Soderstrom KO (2005) Increased proliferation activity measured by immunoreactive Ki67 is associated with survival improvement in rectal/recto sigmoid cancer. World J Gastroenterol 11: 3245-3249. Link: https://goo.gl/IgHvaA
- Reimers MS, Zeestraten EC, van Alphen TC, Dekker JT, Putter H, et al. (2014) Combined analysis of biomarkers of proliferation and apoptosis in colon cancer: an immunohistochemistry-based study using tissue microarray. Int J Colorectal Dis 29: 1043-1052. Link: https://goo.gl/TwF755
- Melling N, Kowitz CM, Simon R, Bokemeyer C, Terracciano L, et al. (2016) High Ki67 expression is an independent good prognostic marker in colorectal cancer. J Clin Pathol 69: 209-214. Link: https://goo.gl/RqPJ9x
- Nishihara K, Shomori K, Fujioka S, Tokuyasu N, Inaba A, et al. (2008) Minichromosome maintenance protein 7 in colorectal cancer: implication of prognostic significance. Int J Oncol 33: 245-252. Link: https://goo.gl/PyMhG2
- Nishihara K, Shomori K, Tamura T, Fujioka S, Ogawa T, et al. (2009) Immunohistochemical expression of geminin in colorectal cancer: Implication of prognostic significance. Oncol rep 21: 1189-1195. Link: https://goo.gl/glynbF
- Hamamoto Y, Shomori K, Nosaka K, Haruki T, Teshima R, et al. (2010) Prognostic significance of Minichromosome maintenance protein 7 and Geminin expression in patients with 109 soft tissue sarcomas. Oncol lett 1: 703-709. Link: https://goo.gl/5vktXU
- Baba Y, Nosho K, Shima K, Irahara N, Kure S, et al. (2009) Aurora-A expression is independently associated with chromosomal instability in colorectal cancer. Neoplasia 11: 418-425. Link: https://goo.gl/zajqV2
- Bravo R, Frank R, Blundell PA, Macdonald-Bravo H (1987) Cyclin/PCNA is the auxiliary protein of DNA polymerase- delta. Nature 326: 515-517. Link: https://goo.gl/kreUeA
- Mulligan JM, Mai KT, Parks W, Gerridzen RG (1987) Proliferating cell nuclear antigen, (PCNA) and MIB 1: Markers of locally advanced and biologically aggressive prostate cancer. Can J Urol 4: 422-425. Link: https://goo.gl/qAI3Kw
- Sethi G, Shanmugam MK, Ramachandran L, Kumar AP, Tergaonkar V (2012) Multifaceted link between cancer and inflammation. Bioscience reports 32: 1-15. Link: https://goo.gl/vbdsNc
- Mager LF, Wasmer M, Rau TT, Krebs P (2016) Cytokine-induced Modulation of Colorectal Cancer. Front Oncol 6. Link: https://goo.gl/wER4kl
- Vaiserman AM, Moskalev AA, Pasyukova EG (2015) Parallels between mammals and flies in Inflammatory Bowel Disease In: Anonymous Life Extension. Springer 151-159. Link: https://goo.gl/BiikKN
- Neu J, Walker WA (2011) Necrotizing enterocolitis. New England Journal of Medicine 364: 255-264. Link: https://goo.gl/4sEqEh
- Patel RM, Denning PW (2015) Intestinal microbiota and its relationship with necrotizing enterocolitis. Pediatr Res 78: 232-238. Link: https://goo.gl/FP4N8l
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R (2004) Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118: 229-241. Link: https://goo.gl/2ClA9d
- Sansonetti P (2011) To be or not to be a pathogen: that is the mucosally relevant question. Mucosal immunol 4: 8-14. Link: https://goo.gl/eF3kqD
- Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, et al. (2011) Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol 17: 1519-1528. Link: https://goo.gl/zoLx9a
- Chen W, Liu F, Ling Z, Tong X, Xiang C (2012) Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS one 7: e39743. Link: https://goo.gl/HJqbG3
- Wang W, Chen L, Zhou R, Wang X, Song L, et al. (2014) Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol 52: 398-406. Link: https://goo.gl/B0Oytv
- Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, et al. (2016) The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome 4: 69. Link: https://goo.gl/cxE5cA
- Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, et al. (2012) The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PloS one 7: e51907. Link: https://goo.gl/YiIWL5
- Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, et al. (2012) Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med 209: 903-911. Link: https://goo.gl/lLQgAm
- Tyler AD, Knox N, Kabakchiev B, Milgrom R, Kirsch R, et al. (2013) Characterization of the gut-associated microbiome in inflammatory pouch complications following ileal pouch-anal anastomosis. PloS one 8: e66934. Link: https://goo.gl/UM8T3b
- Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, et al. (2014) The treatment-naive microbiome in new-onset Crohn’s disease. Cell host microbe 15: 382-392. Link: https://goo.gl/lXoLpX
- Liguori G, Lamas B, Richard ML, Brandi G, Da Costa G, et al. (2016) Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J Crohns Colitis 10: 296-305. Link: https://goo.gl/E0L64m
- Wang T, Cai G, Qiu Y, Fei N, Zhang M, et al. (2012) Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 6: 320-329. Link: https://goo.gl/yLkLMK
- Kumari R, Ahuja V, Paul J (2013) Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of North India. World J Gastroenterol 19: 3404-3414. Link: https://goo.gl/NL6tb6
- Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, et al. (2014) A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63: 1275-1283. Link: https://goo.gl/u3pzpx
- Tilg H, Danese S (2014) Roseburia hominis: a novel guilty player in ulcerative colitis pathogenesis? Gut 63: 1204-1205. Link: https://goo.gl/0sJkIq
- Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, et al. (2011) Towards the human colorectal cancer microbiome. PloS one6: e20447. Link: https://goo.gl/od0N8T
- Geng J, Fan H, Tang X, Zhai H, Zhang Z (2013) Diversified pattern of the human colorectal cancer microbiome. Gut pathog 5: 2. Link: https://goo.gl/buiRHe
- Rafter J (2004) The effects of probiotics on colon cancer development. Nutr Res Rev 17: 277-284. Link: https://goo.gl/w3IJhJ
- Ishikawa H, Akedo I, Otani T, Suzuki T, Nakamura T, et al. (2005) Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. Int J Cancer 116: 762-767. Link: https://goo.gl/Hgzcjz
- Gianotti L, Morelli L, Galbiati F, Rocchetti S, Coppola S, et al. (2010) A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World J Gastroenterol 16: 167-175. Complete. Link: https://goo.gl/6VVxze
- Orlando A, Refolo M, Messa C, Amati L, Lavermicocca P, et al. (2012) Antiproliferative and proapoptotic effects of viable or heat-killed Lactobacillus paracasei IMPC2. 1 and Lactobacillus rhamnosus GG in HGC-27 gastric and DLD-1 colon cell lines. Nutr Cancer 64: 1103-1111. Link: https://goo.gl/KMiB98
- Uccello M, Malaguarnera G, Basile F, D’agata V, Malaguarnera M, et al. (2012) Potential role of probiotics on colorectal cancer prevention. BMC surg 12: S35. Link: https://goo.gl/VeOt61
- Zhong L, Zhang X, Covasa M (2014) Emerging roles of lactic acid bacteria in protection against colorectal cancer. World J Gastroenterol 20: 7878-7886. Link: https://goo.gl/HV9clY
- de Moreno de LeBlanc A, LeBlanc JG (2014) Effect of probiotic administration on the intestinal microbiota, current knowledge and potential applications. World J Gastroenterol 20: 16518-16528. Link: https://goo.gl/iNFHT3
- Lievin-Le Moal V, Servin AL (2014) Anti-infective activities of lactobacillus strains in the human intestinal microbiota: from probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin Microbiol Rev 27: 167-199. Link: https://goo.gl/0tpxco
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, et al. (2013) Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500: 232-236. Link: https://goo.gl/3FUA2l
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, et al. (2006) Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55: 205-211. Link: https://goo.gl/zWM29w
- Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, et al. (2006) Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis 12: 106-111. Link: https://goo.gl/4ooObt
- Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, et al. (2011) Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 60: 631-637. Link: https://goo.gl/XWdceV
- Miquel S, Martin R, Rossi O, Bermudez-Humaran L, Chatel J, et al. (2013) Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol 16: 255-261. Link: https://goo.gl/8lmHJv
- Vigsnaes LK, Van den Abbeele P, Sulek K, Frandsen HL, Steenholdt C, et al. (2013) Microbiotas from UC patients display altered metabolism and reduced ability of LAB to colonize mucus. Sci rep 3: 1110. Link: https://goo.gl/QV23Fe
- Moore WE, Moore LH (1995) Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol 61: 3202-3207. Link: https://goo.gl/PysoZz
- Neurath M (2014) New targets for mucosal healing and therapy in inflammatory bowel diseases. Mucosal Immunol 7: 6-19. Link: https://goo.gl/UkAZjF
- Bangi E, Murgia C, Teague AG, Sansom OJ, Cagan RL (2016) Functional exploration of colorectal cancer genomes using Drosophila. Nature communications 7: 13615. Link: https://goo.gl/6Pmdus
- Dovey H, John V, Anderson J, Chen L, de Saint Andrieu P, et al. (2001) Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem 76: 173-181. Link: https://goo.gl/srRpcu
- Micchelli CA, Esler WP, Kimberly WT, Jack C, Berezovska O, et al. (2003) Gamma-secretase/presenilin inhibitors for Alzheimer's disease phenocopy Notch mutations in Drosophila. FASEB J 17: 79-81. Link: https://goo.gl/fA5EcW
- Mazzoletti M, Bortolin F, Brunelli L, Pastorelli R, Di Giandomenico S, et al. (2011) Combination of PI3K/mTOR inhibitors: antitumor activity and molecular correlates. Cancer research 71: 4573-4584. Link: https://goo.gl/zkxF7H
- Parisi F, Riccardo S, Daniel M, Saqcena M, Kundu N, et al. (2011) Drosophila insulin and target of rapamycin, et al. (TOR) pathways regulate GSK3 beta activity to control Myc stability and determine Myc expression in vivo. BMC biology 9: 65. Link: https://goo.gl/2r73Yv
- Bangi E, Pitsouli C, Rahme LG, Cagan R, Apidianakis Y (2012) Immune response to bacteria induces dissemination of Ras-activated Drosophila hindgut cells. EMBO Rep 13: 569-576. Link: https://goo.gl/TJxQiD
- Shin SC, Kim SH, You H, Kim B, Kim AC, et al. (2011) Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334: 670-674. Link: https://goo.gl/svrkzX
- Golinska E, Tomusiak A, Gosiewski T, Wiecek G, Machul A, et al. (2013) Virulence factors of Enterococcus strains isolated from patients with inflammatory bowel disease. World J Gastroenterol 19: 3562-3572. Link: https://goo.gl/N1fsH8
- Rajilić-Stojanović M, Vos WM (2014) The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38: 996-1047. Link: https://goo.gl/ESukyV
- McLeod CJ, Wang L, Wong C, Jones DL (2010) Stem cell dynamics in response to nutrient availability. Curr Biol 20: 2100-2105. Link: https://goo.gl/nqv8NG
- Deng H, Gerencser AA, Jasper H (2015) Signal integration by Ca2 regulates intestinal stem-cell activity. Nature 528: 212–217. Link: https://goo.gl/lFI6V4
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, et al. (2014) Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13: 397-406. Link: https://goo.gl/PoJ3qt
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, et al. (2014) A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355-364. Link: https://goo.gl/QyCwmM
- Hamada F, Murata Y, Nishida A, Fujita F, Tomoyasu Y, et al. (1999) Identification and characterization of E-APC, a novel Drosophila homologue of the tumour suppressor APC. Genes to Cells 4: 465-474. Link: https://goo.gl/Ha9fp4
- Jin S, Martinek S, Joo WS, Wortman JR, Mirkovic N, et al. (2000) Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc Natl Acad Sci U S A 97: 7301-7306. Link: https://goo.gl/t5Utz5
- Stefancsik R, Sarkar S (2003) Relationship between the DNA binding domains of SMAD and NFI/CTF transcription factors defines a new superfamily of genes. DNA Sequence 14: 233-239. Link: https://goo.gl/BPRX95
- Brock HW (1987) Sequence and genomic structure of ras homologues Dmras85D and Dmras64B of Drosophila melanogaster. Gene 51: 129-137. Link: https://goo.gl/b7SC35
- Gallant P, Shiio Y, Cheng PF, Parkhurst SM, Eisenman RN (1996) Myc and Max homologs in Drosophila. Science 274: 1523. Link: https://goo.gl/h824Og
- Tsuda L, Inoue YH, Yoo M, Mizuno M, Hata M, Lim Y, et al. (1993) A protein kinase similar to MAP kinase activator acts downstream of the raf kinase in Drosophila. Cell 72: 407-414. Link: https://goo.gl/1NDkcr
- Witte HT, Jeibmann A, Klämbt C, Paulus W (2009) Modeling glioma growth and invasion in Drosophila melanogaster. Neoplasia 11: 882-888. Link: https://goo.gl/0nHnEg
- Mark GE, MacIntyre RJ, Digan ME, Ambrosio L, Perrimon N (1987) Drosophila melanogaster homologs of the raf oncogene. Molecular and cellular biology 7: 2134-2140. Link: https://goo.gl/xGlU5m
- Smith A, Smith A, Alrubaie S, Coehlo C, Leevers SJ, et al. (1999) Alternative splicing of the Drosophila PTEN gene. Biochim Biophys Acta 1447: 313-317. Link: https://goo.gl/cL8NOY
- Orian A, Grewal SS, Knoepfler PS, Edgar BA, Parkhurst SM, et al. (2005) Genomic binding and transcriptional regulation by the Drosophila Myc and Mnt transcription factors. Cold Spring Harb Symp Quant Biol 70: 299-307. Link: https://goo.gl/sjBssT
- Stewart S, Guan KL (2000) The dominant negative Ras mutant, N17Ras, can inhibit signaling independently of blocking Ras activation. J Biol Chem 275: 8854-8862. Link: https://goo.gl/INPLft
- Klingensmith J, Noll E, Perrimon N (1989) The segment polarity phenotype of Drosophila involves differential tendencies toward transformation and cell death. Dev biol 134: 130-145. Link: https://goo.gl/C4leV7
- Sander V, Eivers E, Choi RH, De Robertis EM (2010) Drosophila Smad2 opposes Mad signaling during wing vein development. PloS one 5: e10383. Link: https://goo.gl/Iy5URo
- Pickeral OK, Li JZ, Barrow I, Boguski MS, Makałowski W, et al. (2000) Classical oncogenes and tumor suppressor genes: a comparative genomics perspective. Neoplasia 2: 280-286. Link: https://goo.gl/epjJka
- Sekelsky JJ, Brodsky MH, Burtis KC (2000) DNA repair in Drosophila: insights from the Drosophila genome sequence. J Cell Biol 150: F31-6. Link: https://goo.gl/y5eW0V
- Flores C, Engels W (1999) Microsatellite instability in Drosophila spellchecker1 (MutS homolog) mutants. Proc Natl Acad Sci U S A 96: 2964-2969. Link: https://goo.gl/XJ7Fo8
- Klöting N, Follak N, Klöting I (2005) Diabetes per se and metabolic state influence gene expression in tissue-dependent manner of BB/OK rats. Diabetes Metab Res Rev 21: 281-287. Link: https://goo.gl/XyaLA3
- Xi X, Tatei K, Kihara Y, Izumi T (2014) Expression pattern of class I phosphoinositide 3-kinase and distribution of its product, phosphatidylinositol-3, 4, 5-trisphosphate, during Drosophila embryogenesis. Gene Expr Patterns 15: 88-95. Link: https://goo.gl/axS5zT
- Hurvitz JR, Suwairi WM, Van Hul W, El-Shanti H, Superti-Furga A, et al. (1999) Mutations in the CCN gene family member WISP3 cause progressive pseudorheumatoid dysplasia. Nat Genet 23: 94-98. Link: https://goo.gl/1UuLXy
- Childs SR, Wrana JL, Arora K, Attisano L, O'Connor MB, et al. (1993) Identification of a Drosophila activin receptor. Proc Natl Acad Sci U S A 90: 9475-9479. Link: https://goo.gl/PYPQVb
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, et al. (2001) A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292: 107-110. Link: https://goo.gl/Cfi8rb
- Willert K, Logan CY, Arora A, Fish M, Nusse R (1999) A Drosophila Axin homolog, Daxin, inhibits Wnt signaling. Development 126: 4165-4173. Link: https://goo.gl/RQ9gK7
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, et al. (2002) Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome biol 3: research0079. 1. Link: https://goo.gl/7OLxOf
- Fogarty P, Kalpin RF, Sullivan W (1994) The Drosophila maternal-effect mutation grapes causes a metaphase arrest at nuclear cycle. Development 120: 2131-2131. Link: https://goo.gl/LBhPwj
- Gorski MM, Romeijn RJ, Eeken JC, de Jong AW, van Veen BL, et al. (2004) Disruption of Drosophila Rad50 causes pupal lethality, the accumulation of DNA double-strand breaks and the induction of apoptosis in third instar larvae. DNA repair 3: 603-615. Link: https://goo.gl/WNXcBF
- Lipscombe D, Allen SE, Toro CP (2013) Control of neuronal voltage-gated calcium ion channels from RNA to protein. Trends Neurosci 36: 598-609. Link: https://goo.gl/SeUV9z
- Oldham S, Stocker H, Laffargue M, Wittwer F, Wymann M, et al. (2002) The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP (3) levels. Development 129: 4103-4109. Link: https://goo.gl/QDqM74
- Hallgrímsson B, Hall BK (2011) Epigenetics: linking genotype and phenotype in development and evolution. Univ of California Press. Link: https://goo.gl/BjHCty
- Levanon D, Glusman G, Bettoun D, Ben-Asher E, Negreanu V, et al. (2003) Phylogenesis and regulated expression of the RUNT domain transcription factors RUNX1 and RUNX3. Blood Cells Mol Dis 30: 161-163. Link: https://goo.gl/jIAvMM
- Stec W, Vidal O, Zeidler MP (2013) Drosophila SOCS36E negatively regulates JAK/STAT pathway signaling via two separable mechanisms. Mol Biol Cell 24: 3000-3009. Link: https://goo.gl/di0EIJ
- Asha H, Nagy I, Kovacs G, Stetson D, Ando I, et al. (2003) Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163: 203-215. Link: https://goo.gl/ffsRII
- Jung CH, Seo M, Otto NM, Kim D (2011) ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy 7: 1212-1221. Link: https://goo.gl/b99HwZ
- Oppelt A, Lobert VH, Haglund K, Mackey AM, Rameh LE, et al. (2013) Production of phosphatidylinositol 5-phosphate via PIKfyve and MTMR3 regulates cell migration. EMBO reports 14: 57-64. Link: https://goo.gl/Mkk1Up
- Yamazaki Y, Schonherr C, Varshney GK, Dogru M, Hallberg B, et al. (2013) Goliath family E3 ligases regulate the recycling endosome pathway via VAMP3 ubiquitylation. EMBO J 32: 524-537. Link: https://goo.gl/QGjKRM
- Lee S, Liu HP, Lin WY, Guo H, Lu B (2010) LRRK2 kinase regulates synaptic morphology through distinct substrates at the presynaptic and postsynaptic compartments of the Drosophila neuromuscular junction. J Neurosci 30: 16959-16969. Link: https://goo.gl/CQP0oX
- Filippov V, Filippova M, Sehnal F, Gill SS (2000) Temporal and spatial expression of the cell-cycle regulator cul-1 in Drosophila and its stimulation by radiation-induced apoptosis. The Journal of experimental biology 203: 2747-2756. Link: https://goo.gl/BkcMyh
- Hoskins RA, Smith CD, Carlson JW, Carvalho AB, Halpern A, et al. (2002) Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome boil 3: research0085. 1. Link: https://goo.gl/1547Qr
- Imler JL, Bulet P (2005) Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem Immunol Allergy 86: 1-21. Link: https://goo.gl/EeGY2v
- Corwin HO, Hanratty WP (1976) Characterization of a unique lethal tumorous mutation in Drosophila. Mol Gen Genet 144: 345-347. Link: https://goo.gl/QPVVzO
- Yeh TC, Dondi E, Uze G, Pellegrini S (2000) A dual role for the kinase-like domain of the tyrosine kinase Tyk2 in interferon-alpha signaling. Proc Natl Acad Sci U S A 97: 8991-8996. Link: https://goo.gl/2dCkHW
- Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, et al. (1995) Nramp defines a family of membrane proteins. Proc Natl Acad Sci U S A 92: 10089-10093. Link: https://goo.gl/kqRl7w
- Matova N, Anderson KV (2010) Drosophila Rel proteins are central regulators of a robust, multi-organ immune network. J Cell Sci 123: 627-633. Link: https://goo.gl/oEKU2m
- Kramer JM, Davidge JT, Lockyer JM, Staveley BE (2003) Expression of Drosophila FOXO regulates growth and can phenocopy starvation. BMC Dev Biol 3: 5. Link: https://goo.gl/gB1Ern
- Henderson KD, Andrew DJ (1998) Identification of a Novel Drosophila SMAD on the X Chromosome. Biochem Biophys Res Commun 252: 195-201. Link: https://goo.gl/XeYx9I
- Maduzia LL, Padgett RW (1997) Drosophila MAD, a Member of the Smad Family, Translocates to the Nucleus upon Stimulation of the dpp Pathway. Biochem Biophys Res Commun 238: 595-598. Link: https://goo.gl/OjVuM8
- Lawoko-Kerali G, Rivolta MN, Holley M (2002) Expression of the transcription factors GATA3 and Pax2 during development of the mammalian inner ear. J Comp Neurol 442: 378-391. Link: https://goo.gl/PvIwUS
- Wang S, Yin J, Chen D, Nie F, Song X, et al. (2013) Small-molecule modulation of Wnt signaling via modulating the Axin-LRP5/6 interaction. Nature chemical biology 9: 579-585. Link: https://goo.gl/MHUD5h
- Lefevre G, Ratty FJ, Hanks GD (1953) Frequency of Notch Mutations Induced in Normal, Duplicated and Inverted X-Chromosomes of Drosophila Melanogaster. Genetics 38: 345-359. Link: https://goo.gl/EnJUAp
- Regulski M, Tully T (1995) Molecular and biochemical characterization of dNOS: a Drosophila Ca2+/calmodulin-dependent nitric oxide synthase. Proc Natl Acad Sci U S A 92: 9072-9076. Link: https://goo.gl/69h76H
- Kidwell JF (1972) The effective lethal phase of the curly mutant in Drosophila melanogaster. J Hered 63: 100. Link: https://goo.gl/8IDjpI
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, et al. (2002) The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome biol 3: research 0084. 1. Link: https://goo.gl/dnqC7t
- Taylor CA, Stanley KN, Shirras AD (1997) The Orct gene of Drosophila melanogaster codes for a putative organic cation transporter with six or 12 transmembrane domains. Gene 201: 69-74. Link: https://goo.gl/scCOaB
- Missirlis F, Rahlfs S, Dimopoulos N, Bauer H, Becker K, et al. (2003) A putative glutathione peroxidase of Drosophila encodes a thioredoxin peroxidase that provides resistance against oxidative stress but fails to complement a lack of catalase activity. Biol Chem 384: 463472. Link: https://goo.gl/0inlhf
- Kim RH, Peters M, Jang Y, Shi W, Pintilie M, et al. (2005) DJ1, a novel regulator of the tumor suppressor PTEN. Cancer cell 7: 263-273. Link: https://goo.gl/tUAQ0G
- Vassin H, Campos-Ortega JA (1987) Genetic Analysis of Delta, a Neurogenic Gene of Drosophila melanogaster. Genetics 116: 433-445. Link: https://goo.gl/wV2b3X
- Arnold RS, Shi J, Murad E, Whalen AM, Sun CQ, Polavarapu R, et al. (2001) Hydrogen peroxide mediates the cell growth and transformation caused by the mitogenic oxidase Nox1. Proc Natl Acad Sci U S A 98: 5550-5555. Link: https://goo.gl/edD2aM
- Syed ZA, Härd T, Uv A, van Dijk-Härd IF (2008) A potential role for Drosophila mucins in development and physiology. PLoS One 3: e3041. Link: https://goo.gl/H2x29Q
- Barry WE, Thummel CS (2016) The Drosophila HNF4 nuclear receptor promotes glucose-stimulated insulin secretion and mitochondrial function in adults. eLife 5: 10.7554/eLife.11183. Link: https://goo.gl/mDWCQi
- Jacobs HW, Richter DO, Venkatesh TR, Lehner CF (2002) Completion of mitosis requires neither fzr/rap nor fzr2, a male germline-specific Drosophila Cdh1 homolog. Current biology 12: 1435-1441. Link: https://goo.gl/rok6zo
- Obregón FP, Papalardo C, Castro S, Guerberoff G, Cantera R (2015) Putative synaptic genes defined from a Drosophila whole body developmental transcriptome by a machine learning approach. BMC genomics 16: 694. Link: https://goo.gl/l2UYDG
- Urbano JM, Torgler CN, Molnar C, Tepass U, Lopez-Varea A, et al. (2009) Drosophila laminins act as key regulators of basement membrane assembly and morphogenesis. Development 136: 4165-4176. Link: https://goo.gl/yhEXrH
- Yan M, Ha JH, Dhanasekaran DN (2015) Gα13 Stimulates the Tyrosine Phosphorylation of Ric-8A. J Mol Signal 10: 3. Link: https://goo.gl/Xs5XwL
- Hill E, Broadbent ID, Chothia C, Pettitt J (2001) Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster. J Mol Biol 305: 1011-1024. Link: https://goo.gl/Z6NDgN
- Wei S, Xie Z, Filenova E, Brew K (2003) Drosophila TIMP is a potent inhibitor of MMPs and TACE: similarities in structure and function to TIMP-3. Biochemistry 42: 12200-12207. Link: https://goo.gl/8hNXAd
- Llano E, Pendas AM, Aza-Blanc P, Kornberg TB, Lopez-Otin C (2000) Dm1-MMP, a matrix metalloproteinase from Drosophila with a potential role in extracellular matrix remodeling during neural development. J Biol Chem 275: 35978-35985. Link: https://goo.gl/XiSKNF
- Huang JH, Rajkovic A, Szafranski P, Ochsner S, Richards J, et al. (2003) Expression of Drosophila neoplastic tumor suppressor genes discslarge, scribble, and lethal giant larvae in the mammalian ovary. Gene Expr Patterns 3: 3-11. Link: https://goo.gl/J8l42C
- Takano-Ohmuro H, Takahashi S, Hirose G, Maruyama K (1990) Phosphorylated and dephosphorylated myosin light chains of Drosophila fly and larva. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry 95: 171-177. https://goo.gl/Fcv8lk
- Bender JS, Kinyon JM, Kariyawasam S, Halbur PG, Opriessnig T (2009) Comparison of conventional direct and enrichment culture methods for Erysipelothrix spp. from experimentally and naturally infected swine. J Vet Diagn Invest 21: 863-868. Link: https://goo.gl/MlebwZ









Table 1:
Genomic Instability and Epigenetic Instability related genes and their homology in human, mice and flies.
PROCESS
HUMAN
SNPs/GENES
[136]
MOUSE
SNPs/GENES
[137]
FLY GENES
HOMOLOGY
Chromosomal
Instability
APC
APC
APC-like [138]
Human-mouse: 90%
TP53
Trp53
p53 [139]
Human-mouse: 77%
SMAD4
Smad4
Med [140]
Human-mouse: 98% Human-fly: 78%
KRAS
Kras
Ras85D [141]
Human-mouse: 96% Human-fly: 85%
MYC
Myc
dMyc [142]
Human-mouse: 87%
RAF1
Raf1
D-raf [143]
Human-mouse: 98%
PIK3CA
Pik3ca
PI3K [144]
Human-mouse: 99%
BRAF
Braf
Raf [145]
Human-mouse: 86% Human-fly: 45%
PTEN
Pten
Pten [146]
Human-mouse: 99% Human-fly: 47%
TCF7L2
Fbxw7
Archipelago [147]
Human-mouse: 95%
TCF7L2
Tcf7l2
dTCF [59]
Human-mouse: 97%
NRAS
Nras
NRas [148]
Human-mouse: 99%
AMER1
Amer1
Human-mouse: 77%
CTNNB1
Ctnnb1
arm [149]
Human-mouse: 99% Human-fly: 67%
SMAD2
Smad2
Smad2 [150]
Human-mouse: 99%
Microsatellite Instability
MLH1
Mlh1
Mlh1 [151]
Human-mouse: 88% Human-fly: 58%
MLH3
Mlh3
Human-mouse: 70%
PMS1
Pms1
Human-mouse: 75%
PMS2
Pms2
Pms2 [152]
Human-mouse: 77%
MSH2
Msh2
spel1 [153]
Human-mouse: 93% Human-fly: 45%
MSH3
Msh3
Human-mouse: 82%
MSH6
Msh6
Msh6 [152]
Human-mouse: 85% Human-fly: 44%
Exo1
Exo1
Human-mouse: 73%
TGFB1
Tgfb1
Dlk1/Pref-1 [154]
Human-mouse: 90%
PIK3R1
Pik3r1
Pi3K21B [155]
Human-mouse: 96%
TCF-4
Tcf4
dTCF [59]
Human-mouse: 96%
WISP3
Wisp3
Ccn [156]
Human-mouse: 79%
ACVR2A
Acvr2a
put [157]
Human-mouse: 99% Human-fly: 47%
IGF2R
Igf2r
DInR [158]
Human-mouse: 81%
AXIN2
Axin2
Daxin [159]
Human-mouse: 88%
CDX
Cdx1
Human-mouse: 84%
BAX
Bax
Human-mouse: 92%
PRDM2
Prdm2
Human-mouse: 81%
BCL10
Bcl10
Human-mouse: 91%
PA2G4
Pa2g4
CG10576 [160]
Human-mouse: 99% Human-fly: 56%
FAS
Fas
Human-mouse: 49%
MBD4
Mbd4
Human-mouse: 96%
BLM
Blm
Human-mouse: 75%
CHEK1
Chek1
grp [161]
Human-mouse: 93% Human-fly: 47%
RAD50
Rad50
rad50 [162]
Human-mouse: 92% Human-fly: 29%
Epigenomic
Instability
CACNA1G
Cacna1g
Ca-α1T [163]
Human-mouse: 94%
IGF2
Igf2
Igf [164]
Human-mouse: 82%
NEUROG1
Neurog1
Atonal [165]
Human-mouse: 77%
RUNX3
Runx3
Runt [166]
Human-mouse: 89%
SOCS1
Socs1
SOCS36E [167]
Human-mouse: 92%